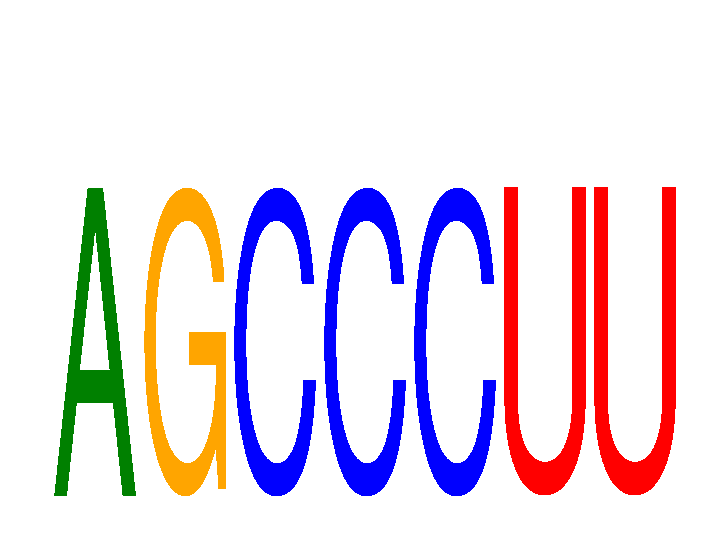Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for AGCCCUU
Z-value: 0.35
miRNA associated with seed AGCCCUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-129-1-3p
|
MIMAT0016994 |
|
mmu-miR-129-2-3p
|
MIMAT0000544 |
Activity profile of AGCCCUU motif
Sorted Z-values of AGCCCUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_3280401 | 0.24 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr12_-_54250646 | 0.22 |
ENSMUST00000039516.4
|
Egln3
|
egl-9 family hypoxia-inducible factor 3 |
| chr13_-_29137673 | 0.22 |
ENSMUST00000067230.6
|
Sox4
|
SRY (sex determining region Y)-box 4 |
| chr11_-_87878301 | 0.15 |
ENSMUST00000020775.9
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr17_-_74601769 | 0.13 |
ENSMUST00000078459.8
ENSMUST00000232989.2 |
Memo1
|
mediator of cell motility 1 |
| chr13_+_94194269 | 0.12 |
ENSMUST00000054274.8
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr4_+_97665843 | 0.12 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr1_+_191449946 | 0.12 |
ENSMUST00000133076.7
ENSMUST00000110855.8 |
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr10_-_117681864 | 0.11 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr17_-_56447332 | 0.10 |
ENSMUST00000001256.11
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr9_-_115139489 | 0.10 |
ENSMUST00000035010.10
|
Stt3b
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) |
| chr12_+_102724223 | 0.10 |
ENSMUST00000046404.8
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr12_-_65010124 | 0.10 |
ENSMUST00000021331.9
|
Klhl28
|
kelch-like 28 |
| chr8_+_111760521 | 0.10 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr10_+_128641520 | 0.10 |
ENSMUST00000219508.2
ENSMUST00000026410.2 |
Dnajc14
|
DnaJ heat shock protein family (Hsp40) member C14 |
| chr4_+_129030710 | 0.10 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr1_+_155034452 | 0.09 |
ENSMUST00000027743.13
ENSMUST00000195302.6 |
Stx6
|
syntaxin 6 |
| chr2_-_131021905 | 0.09 |
ENSMUST00000089510.5
|
Cenpb
|
centromere protein B |
| chr11_-_106107132 | 0.09 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr5_-_21087023 | 0.09 |
ENSMUST00000118174.8
|
Phtf2
|
putative homeodomain transcription factor 2 |
| chr6_+_95094721 | 0.09 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr13_+_120151982 | 0.08 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr5_-_23988696 | 0.08 |
ENSMUST00000119946.8
|
Pus7
|
pseudouridylate synthase 7 |
| chr1_-_170042947 | 0.08 |
ENSMUST00000027979.14
ENSMUST00000123399.2 |
Uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr1_-_165462020 | 0.08 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chr4_-_41713490 | 0.08 |
ENSMUST00000038434.4
|
Rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr14_+_48683797 | 0.08 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chr8_+_10027707 | 0.07 |
ENSMUST00000139793.8
ENSMUST00000048216.6 |
Abhd13
|
abhydrolase domain containing 13 |
| chr5_-_149559667 | 0.07 |
ENSMUST00000074846.14
|
Hsph1
|
heat shock 105kDa/110kDa protein 1 |
| chr11_-_120524362 | 0.07 |
ENSMUST00000058162.14
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr1_-_171854818 | 0.07 |
ENSMUST00000138714.2
ENSMUST00000027837.13 ENSMUST00000111264.8 |
Vangl2
|
VANGL planar cell polarity 2 |
| chr4_+_15265798 | 0.07 |
ENSMUST00000062684.9
|
Tmem64
|
transmembrane protein 64 |
| chr11_-_115918784 | 0.07 |
ENSMUST00000106454.8
|
H3f3b
|
H3.3 histone B |
| chr2_-_24653059 | 0.07 |
ENSMUST00000100348.10
ENSMUST00000041342.12 ENSMUST00000114447.8 ENSMUST00000102939.9 ENSMUST00000070864.14 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr11_-_49603501 | 0.07 |
ENSMUST00000020624.7
ENSMUST00000145353.8 |
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr15_-_31531122 | 0.06 |
ENSMUST00000090227.6
|
Marchf6
|
membrane associated ring-CH-type finger 6 |
| chr13_+_119565424 | 0.06 |
ENSMUST00000026520.14
|
Paip1
|
polyadenylate binding protein-interacting protein 1 |
| chr2_+_121859025 | 0.06 |
ENSMUST00000028668.8
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chr12_-_54842488 | 0.06 |
ENSMUST00000005798.9
|
Snx6
|
sorting nexin 6 |
| chr1_+_180396478 | 0.06 |
ENSMUST00000027777.12
|
Parp1
|
poly (ADP-ribose) polymerase family, member 1 |
| chr10_+_44943262 | 0.06 |
ENSMUST00000099858.4
|
Prep
|
prolyl endopeptidase |
| chr12_-_102844537 | 0.06 |
ENSMUST00000045652.8
ENSMUST00000223554.2 |
Btbd7
|
BTB (POZ) domain containing 7 |
| chr14_-_47059694 | 0.06 |
ENSMUST00000111817.8
ENSMUST00000079314.12 |
Gmfb
|
glia maturation factor, beta |
| chr11_+_30835358 | 0.06 |
ENSMUST00000109430.2
ENSMUST00000203878.3 |
Gpr75
Asb3
|
G protein-coupled receptor 75 ankyrin repeat and SOCS box-containing 3 |
| chr5_-_73805063 | 0.06 |
ENSMUST00000081170.9
|
Sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr1_+_93682621 | 0.06 |
ENSMUST00000027502.16
|
Atg4b
|
autophagy related 4B, cysteine peptidase |
| chr1_-_95595280 | 0.05 |
ENSMUST00000043336.11
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr1_-_157240138 | 0.05 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr13_-_30170031 | 0.05 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr11_-_78056347 | 0.05 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr19_-_41252370 | 0.05 |
ENSMUST00000237871.2
ENSMUST00000025989.10 |
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr8_-_125675901 | 0.05 |
ENSMUST00000034469.7
|
Egln1
|
egl-9 family hypoxia-inducible factor 1 |
| chr11_+_5905693 | 0.05 |
ENSMUST00000002818.9
|
Ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr17_-_66079681 | 0.05 |
ENSMUST00000070673.9
|
Rab31
|
RAB31, member RAS oncogene family |
| chr5_-_67585137 | 0.05 |
ENSMUST00000169190.5
|
Bend4
|
BEN domain containing 4 |
| chr3_+_151916059 | 0.05 |
ENSMUST00000166984.8
|
Fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr1_+_184936306 | 0.05 |
ENSMUST00000194740.6
ENSMUST00000069652.8 |
Rab3gap2
|
RAB3 GTPase activating protein subunit 2 |
| chr7_+_109721230 | 0.05 |
ENSMUST00000033326.10
|
Wee1
|
WEE 1 homolog 1 (S. pombe) |
| chr18_+_36414122 | 0.05 |
ENSMUST00000051301.6
|
Pura
|
purine rich element binding protein A |
| chr7_+_118311740 | 0.04 |
ENSMUST00000106557.8
|
Ccp110
|
centriolar coiled coil protein 110 |
| chr2_+_75489596 | 0.04 |
ENSMUST00000111964.8
ENSMUST00000111962.8 ENSMUST00000111961.8 ENSMUST00000164947.9 ENSMUST00000090792.11 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr18_-_39623698 | 0.04 |
ENSMUST00000115567.8
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chrX_+_74557905 | 0.04 |
ENSMUST00000114070.10
ENSMUST00000033540.6 |
Vbp1
|
von Hippel-Lindau binding protein 1 |
| chr10_-_118705029 | 0.04 |
ENSMUST00000004281.10
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr10_-_94780695 | 0.04 |
ENSMUST00000099337.5
|
Plxnc1
|
plexin C1 |
| chr1_-_131066004 | 0.04 |
ENSMUST00000016670.9
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr12_-_85871281 | 0.04 |
ENSMUST00000021676.12
ENSMUST00000142331.2 |
Erg28
|
ergosterol biosynthesis 28 |
| chr5_+_122988111 | 0.04 |
ENSMUST00000031434.8
ENSMUST00000198602.2 |
Rnf34
|
ring finger protein 34 |
| chr8_-_87611849 | 0.04 |
ENSMUST00000034074.8
|
N4bp1
|
NEDD4 binding protein 1 |
| chr4_+_148225139 | 0.04 |
ENSMUST00000140049.8
ENSMUST00000105707.2 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr4_-_88640691 | 0.04 |
ENSMUST00000094993.3
|
Klhl9
|
kelch-like 9 |
| chr9_+_96078340 | 0.04 |
ENSMUST00000034982.16
ENSMUST00000188008.7 ENSMUST00000188750.7 ENSMUST00000185644.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr3_-_69034425 | 0.04 |
ENSMUST00000194558.6
ENSMUST00000029353.9 |
Kpna4
|
karyopherin (importin) alpha 4 |
| chr1_-_39616445 | 0.04 |
ENSMUST00000062525.11
|
Rnf149
|
ring finger protein 149 |
| chr13_-_101904662 | 0.04 |
ENSMUST00000055518.13
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr18_+_34910064 | 0.03 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr4_+_120711974 | 0.03 |
ENSMUST00000071093.9
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr7_-_125995884 | 0.03 |
ENSMUST00000075671.5
|
Nfatc2ip
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 interacting protein |
| chr3_-_9675130 | 0.03 |
ENSMUST00000041124.13
|
Zfp704
|
zinc finger protein 704 |
| chr1_+_58432021 | 0.03 |
ENSMUST00000050552.15
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr16_+_21828223 | 0.03 |
ENSMUST00000023561.8
|
Senp2
|
SUMO/sentrin specific peptidase 2 |
| chr1_-_192812509 | 0.03 |
ENSMUST00000085555.7
|
Utp25
|
UTP25 small subunit processome component |
| chr2_-_6889783 | 0.03 |
ENSMUST00000170438.8
ENSMUST00000114924.10 ENSMUST00000114934.11 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr1_+_92759324 | 0.03 |
ENSMUST00000045970.8
|
Gpc1
|
glypican 1 |
| chr3_-_37180093 | 0.03 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr9_-_43017249 | 0.03 |
ENSMUST00000165665.9
|
Arhgef12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr4_+_45342069 | 0.03 |
ENSMUST00000155551.8
|
Dcaf10
|
DDB1 and CUL4 associated factor 10 |
| chr14_-_66361931 | 0.03 |
ENSMUST00000070515.2
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr16_+_64672334 | 0.03 |
ENSMUST00000067744.8
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr1_-_181039509 | 0.03 |
ENSMUST00000162819.9
ENSMUST00000237749.2 |
Wdr26
|
WD repeat domain 26 |
| chr18_+_24087725 | 0.03 |
ENSMUST00000225682.2
ENSMUST00000060762.6 |
Zfp397
|
zinc finger protein 397 |
| chr1_+_84817547 | 0.02 |
ENSMUST00000097672.4
|
Fbxo36
|
F-box protein 36 |
| chr9_-_82857528 | 0.02 |
ENSMUST00000034787.12
|
Phip
|
pleckstrin homology domain interacting protein |
| chr19_-_4319170 | 0.02 |
ENSMUST00000037992.16
ENSMUST00000113852.6 ENSMUST00000236794.2 |
Ssh3
|
slingshot protein phosphatase 3 |
| chr12_-_51876282 | 0.02 |
ENSMUST00000042052.9
ENSMUST00000179265.8 |
Hectd1
|
HECT domain E3 ubiquitin protein ligase 1 |
| chr9_-_117081518 | 0.02 |
ENSMUST00000111773.10
ENSMUST00000068962.14 ENSMUST00000044901.14 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr9_+_45749869 | 0.02 |
ENSMUST00000078111.11
ENSMUST00000034591.11 |
Bace1
|
beta-site APP cleaving enzyme 1 |
| chr15_+_99615396 | 0.02 |
ENSMUST00000023760.13
ENSMUST00000162194.2 |
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr16_-_31898088 | 0.02 |
ENSMUST00000023467.9
|
Pak2
|
p21 (RAC1) activated kinase 2 |
| chr13_-_104246084 | 0.02 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr3_+_52948938 | 0.02 |
ENSMUST00000059562.14
ENSMUST00000147139.2 |
Lhfp
|
lipoma HMGIC fusion partner |
| chr2_-_124965537 | 0.02 |
ENSMUST00000142718.8
ENSMUST00000152367.8 ENSMUST00000067780.10 ENSMUST00000147105.8 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr2_+_163916042 | 0.02 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr18_-_46658957 | 0.02 |
ENSMUST00000036226.6
|
Fem1c
|
fem 1 homolog c |
| chr7_-_118183892 | 0.02 |
ENSMUST00000044195.6
|
Tmc7
|
transmembrane channel-like gene family 7 |
| chr1_-_178165223 | 0.02 |
ENSMUST00000037748.9
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U |
| chr11_+_117006020 | 0.02 |
ENSMUST00000103026.10
ENSMUST00000090433.6 |
Sec14l1
|
SEC14-like lipid binding 1 |
| chr2_+_75662511 | 0.02 |
ENSMUST00000047232.14
ENSMUST00000111952.9 |
Agps
|
alkylglycerone phosphate synthase |
| chr1_+_139349912 | 0.02 |
ENSMUST00000200243.5
ENSMUST00000039867.10 |
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr9_-_114762986 | 0.02 |
ENSMUST00000146623.8
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr6_+_115830431 | 0.02 |
ENSMUST00000112925.8
ENSMUST00000038234.13 ENSMUST00000112923.7 |
Ift122
|
intraflagellar transport 122 |
| chr11_+_78213791 | 0.02 |
ENSMUST00000017534.15
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr18_+_44960813 | 0.02 |
ENSMUST00000037763.11
|
Ythdc2
|
YTH domain containing 2 |
| chr13_-_36918424 | 0.02 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
| chrX_+_135950334 | 0.02 |
ENSMUST00000047852.8
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr5_-_98178811 | 0.02 |
ENSMUST00000031281.14
|
Antxr2
|
anthrax toxin receptor 2 |
| chr15_-_79718423 | 0.02 |
ENSMUST00000109623.8
ENSMUST00000109625.8 ENSMUST00000023060.13 ENSMUST00000089299.6 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr5_+_101912939 | 0.02 |
ENSMUST00000031273.9
|
Cds1
|
CDP-diacylglycerol synthase 1 |
| chr10_+_128212953 | 0.02 |
ENSMUST00000014642.10
|
Ankrd52
|
ankyrin repeat domain 52 |
| chr19_+_21755689 | 0.02 |
ENSMUST00000096194.9
ENSMUST00000025663.8 |
Cemip2
|
cell migration inducing hyaluronidase 2 |
| chr3_+_14643669 | 0.01 |
ENSMUST00000029069.13
ENSMUST00000165922.3 |
E2f5
|
E2F transcription factor 5 |
| chr1_+_74545203 | 0.01 |
ENSMUST00000087215.7
|
Cnot9
|
CCR4-NOT transcription complex, subunit 9 |
| chr8_-_54983027 | 0.01 |
ENSMUST00000067476.9
|
Spcs3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr2_+_20742115 | 0.01 |
ENSMUST00000114606.8
ENSMUST00000114608.3 |
Etl4
|
enhancer trap locus 4 |
| chr5_-_108134869 | 0.01 |
ENSMUST00000145239.2
ENSMUST00000031198.11 |
Dipk1a
|
divergent protein kinase domain 1A |
| chr14_+_73379930 | 0.01 |
ENSMUST00000170370.8
ENSMUST00000164822.8 ENSMUST00000165429.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr18_-_72484126 | 0.01 |
ENSMUST00000114943.11
|
Dcc
|
deleted in colorectal carcinoma |
| chr15_+_59520199 | 0.01 |
ENSMUST00000067543.8
|
Trib1
|
tribbles pseudokinase 1 |
| chr17_+_66090336 | 0.01 |
ENSMUST00000073104.11
ENSMUST00000160664.8 ENSMUST00000162272.2 |
Ppp4r1
|
protein phosphatase 4, regulatory subunit 1 |
| chr7_+_19197192 | 0.01 |
ENSMUST00000137613.9
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr8_-_95602952 | 0.01 |
ENSMUST00000046461.9
|
Dok4
|
docking protein 4 |
| chr11_+_110289941 | 0.01 |
ENSMUST00000020949.12
ENSMUST00000100260.2 |
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr5_-_96309849 | 0.01 |
ENSMUST00000155901.8
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chrX_+_149981074 | 0.01 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr10_-_110845860 | 0.01 |
ENSMUST00000041723.15
|
Zdhhc17
|
zinc finger, DHHC domain containing 17 |
| chr3_-_153650269 | 0.01 |
ENSMUST00000072697.13
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr5_+_73071607 | 0.01 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr12_+_65083093 | 0.01 |
ENSMUST00000120580.8
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr9_+_58536386 | 0.01 |
ENSMUST00000176250.2
|
Nptn
|
neuroplastin |
| chr11_+_89921121 | 0.01 |
ENSMUST00000092788.4
|
Tmem100
|
transmembrane protein 100 |
| chr6_-_32565127 | 0.01 |
ENSMUST00000115096.4
|
Plxna4
|
plexin A4 |
| chr2_+_118998235 | 0.01 |
ENSMUST00000057454.4
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr8_+_124380639 | 0.01 |
ENSMUST00000045487.4
|
Rhou
|
ras homolog family member U |
| chr11_+_85202058 | 0.01 |
ENSMUST00000020835.16
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr3_+_125197722 | 0.01 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr2_+_26899935 | 0.01 |
ENSMUST00000114005.9
ENSMUST00000114004.8 ENSMUST00000114006.8 ENSMUST00000114007.8 ENSMUST00000133807.2 |
Cacfd1
|
calcium channel flower domain containing 1 |
| chr13_+_55357585 | 0.01 |
ENSMUST00000224973.2
ENSMUST00000099490.3 |
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr18_+_44513547 | 0.01 |
ENSMUST00000202306.2
ENSMUST00000025350.10 |
Dcp2
|
decapping mRNA 2 |
| chr7_-_81104423 | 0.00 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr2_+_30282266 | 0.00 |
ENSMUST00000028209.15
|
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr6_+_113355076 | 0.00 |
ENSMUST00000156898.5
ENSMUST00000203578.3 ENSMUST00000171058.8 |
Arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr3_+_88439616 | 0.00 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr2_-_167032068 | 0.00 |
ENSMUST00000059826.10
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr2_+_83642910 | 0.00 |
ENSMUST00000051454.4
|
Fam171b
|
family with sequence similarity 171, member B |
| chr2_-_37537224 | 0.00 |
ENSMUST00000028279.10
|
Strbp
|
spermatid perinuclear RNA binding protein |
| chr16_+_76810588 | 0.00 |
ENSMUST00000239066.2
ENSMUST00000023580.8 |
Usp25
|
ubiquitin specific peptidase 25 |
| chr8_-_9821021 | 0.00 |
ENSMUST00000208933.2
ENSMUST00000110969.5 |
Fam155a
|
family with sequence similarity 155, member A |
| chr2_+_157266175 | 0.00 |
ENSMUST00000029175.14
ENSMUST00000092576.11 |
Src
|
Rous sarcoma oncogene |
| chr4_+_138181616 | 0.00 |
ENSMUST00000050918.4
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chrX_-_103714653 | 0.00 |
ENSMUST00000042070.6
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr5_-_92110927 | 0.00 |
ENSMUST00000031345.15
|
Rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr19_+_46385321 | 0.00 |
ENSMUST00000039922.13
ENSMUST00000111867.9 ENSMUST00000120778.8 |
Sufu
|
SUFU negative regulator of hedgehog signaling |
| chr12_+_64964674 | 0.00 |
ENSMUST00000058135.6
ENSMUST00000220993.2 |
Gm527
|
predicted gene 527 |
| chr2_-_25982160 | 0.00 |
ENSMUST00000114159.9
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr10_+_69048464 | 0.00 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr2_-_52566583 | 0.00 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr18_+_31892933 | 0.00 |
ENSMUST00000115808.4
|
Ammecr1l
|
AMME chromosomal region gene 1-like |
| chr7_-_81356653 | 0.00 |
ENSMUST00000026922.15
|
Homer2
|
homer scaffolding protein 2 |
| chr11_-_66416790 | 0.00 |
ENSMUST00000066679.7
|
Shisa6
|
shisa family member 6 |
| chr15_-_98676007 | 0.00 |
ENSMUST00000226655.2
ENSMUST00000228546.2 ENSMUST00000023732.12 ENSMUST00000226610.2 |
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr18_-_31580436 | 0.00 |
ENSMUST00000025110.5
|
Syt4
|
synaptotagmin IV |
| chr5_+_17779273 | 0.00 |
ENSMUST00000030568.14
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr19_+_16110176 | 0.00 |
ENSMUST00000025541.6
|
Gnaq
|
guanine nucleotide binding protein, alpha q polypeptide |
| chr2_+_52747855 | 0.00 |
ENSMUST00000155586.9
ENSMUST00000090952.11 ENSMUST00000127122.9 ENSMUST00000049483.14 ENSMUST00000050719.13 |
Fmnl2
|
formin-like 2 |
| chr7_+_18828519 | 0.00 |
ENSMUST00000049454.6
|
Six5
|
sine oculis-related homeobox 5 |
| chr1_+_191058109 | 0.00 |
ENSMUST00000027940.6
|
Pacc1
|
proton activated chloride channel 1 |
| chr18_-_61859373 | 0.00 |
ENSMUST00000062991.9
ENSMUST00000238010.2 |
Grpel2
|
GrpE-like 2, mitochondrial |
| chr7_+_63937413 | 0.00 |
ENSMUST00000032736.11
|
Mtmr10
|
myotubularin related protein 10 |
| chr8_-_74080101 | 0.00 |
ENSMUST00000119826.7
ENSMUST00000212459.2 |
Large1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr6_+_36364990 | 0.00 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr8_+_108020092 | 0.00 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr1_+_152275575 | 0.00 |
ENSMUST00000044311.9
|
Colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr15_-_31367668 | 0.00 |
ENSMUST00000110410.10
ENSMUST00000076942.5 |
Ankrd33b
|
ankyrin repeat domain 33B |
| chr5_+_32768515 | 0.00 |
ENSMUST00000202543.4
ENSMUST00000072311.13 |
Yes1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr4_-_35845204 | 0.00 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr5_+_67417908 | 0.00 |
ENSMUST00000037918.12
ENSMUST00000162543.8 ENSMUST00000161233.8 ENSMUST00000160352.8 |
Tmem33
|
transmembrane protein 33 |
| chr7_-_141925947 | 0.00 |
ENSMUST00000084412.6
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr16_-_9812410 | 0.00 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
Network of associatons between targets according to the STRING database.
First level regulatory network of AGCCCUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 | 0.1 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.0 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.0 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |