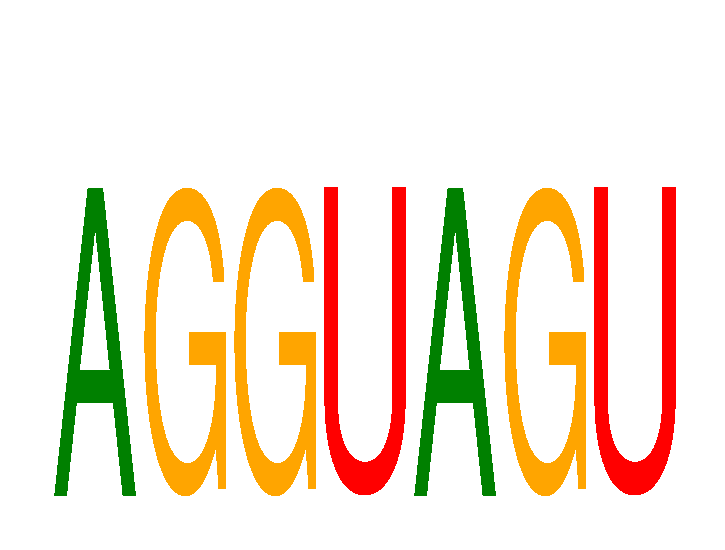Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for AGGUAGU
Z-value: 0.40
miRNA associated with seed AGGUAGU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-196a-5p
|
MIMAT0000518 |
|
mmu-miR-196b-5p
|
MIMAT0001081 |
Activity profile of AGGUAGU motif
Sorted Z-values of AGGUAGU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_95855442 | 0.23 |
ENSMUST00000030306.14
|
Hook1
|
hook microtubule tethering protein 1 |
| chr4_+_140427799 | 0.21 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chrX_+_35592006 | 0.18 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr2_+_152907875 | 0.18 |
ENSMUST00000238488.2
ENSMUST00000129377.8 ENSMUST00000109800.2 |
Ccm2l
|
cerebral cavernous malformation 2-like |
| chr9_+_51958453 | 0.17 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr4_-_133746138 | 0.17 |
ENSMUST00000051674.3
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr12_+_100165694 | 0.16 |
ENSMUST00000110082.11
|
Calm1
|
calmodulin 1 |
| chr6_-_52203146 | 0.15 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr9_-_103242096 | 0.15 |
ENSMUST00000116517.9
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr11_-_76737794 | 0.15 |
ENSMUST00000021201.6
|
Cpd
|
carboxypeptidase D |
| chr7_-_16657825 | 0.14 |
ENSMUST00000019514.10
|
Calm3
|
calmodulin 3 |
| chr4_+_6365650 | 0.13 |
ENSMUST00000029912.11
ENSMUST00000103008.12 |
Sdcbp
|
syndecan binding protein |
| chr17_-_26314461 | 0.13 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr15_-_76422928 | 0.12 |
ENSMUST00000023219.9
|
Fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr4_+_54947976 | 0.12 |
ENSMUST00000098070.10
|
Zfp462
|
zinc finger protein 462 |
| chr11_+_57692399 | 0.11 |
ENSMUST00000020826.6
|
Sap30l
|
SAP30-like |
| chr5_-_124233812 | 0.11 |
ENSMUST00000031354.11
|
Abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr14_+_30437556 | 0.11 |
ENSMUST00000054230.12
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr6_-_49191891 | 0.11 |
ENSMUST00000031838.9
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr7_+_120442048 | 0.11 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr1_-_37580084 | 0.10 |
ENSMUST00000151952.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr13_-_8921732 | 0.10 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr2_+_36026395 | 0.10 |
ENSMUST00000028250.9
|
Mrrf
|
mitochondrial ribosome recycling factor |
| chr2_-_152239966 | 0.09 |
ENSMUST00000063332.9
ENSMUST00000182625.2 |
Sox12
|
SRY (sex determining region Y)-box 12 |
| chr3_-_10505113 | 0.09 |
ENSMUST00000029047.12
ENSMUST00000195822.2 ENSMUST00000099223.11 |
Snx16
|
sorting nexin 16 |
| chr8_-_84831391 | 0.09 |
ENSMUST00000041367.9
ENSMUST00000210279.2 |
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr6_-_52181393 | 0.09 |
ENSMUST00000048794.7
|
Hoxa5
|
homeobox A5 |
| chr4_+_28813125 | 0.09 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr12_+_108758871 | 0.08 |
ENSMUST00000021692.9
|
Yy1
|
YY1 transcription factor |
| chr12_-_31401432 | 0.08 |
ENSMUST00000110857.5
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr8_-_47805383 | 0.07 |
ENSMUST00000110367.10
|
Stox2
|
storkhead box 2 |
| chr2_+_31135813 | 0.07 |
ENSMUST00000000199.8
|
Ncs1
|
neuronal calcium sensor 1 |
| chr6_+_54572096 | 0.07 |
ENSMUST00000119706.8
|
Plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr13_-_38178059 | 0.06 |
ENSMUST00000225319.2
ENSMUST00000225246.2 ENSMUST00000021864.8 |
Ssr1
|
signal sequence receptor, alpha |
| chr1_+_93731662 | 0.06 |
ENSMUST00000027505.13
|
Ing5
|
inhibitor of growth family, member 5 |
| chr17_+_27775613 | 0.06 |
ENSMUST00000231780.2
ENSMUST00000232253.2 ENSMUST00000232552.2 ENSMUST00000117600.9 |
Hmga1
|
high mobility group AT-hook 1 |
| chr18_+_31742565 | 0.06 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr18_+_12261798 | 0.05 |
ENSMUST00000025270.8
|
Riok3
|
RIO kinase 3 |
| chr2_+_60040231 | 0.05 |
ENSMUST00000102748.11
|
Marchf7
|
membrane associated ring-CH-type finger 7 |
| chr2_+_167922924 | 0.05 |
ENSMUST00000052125.7
|
Pard6b
|
par-6 family cell polarity regulator beta |
| chr13_-_117162041 | 0.05 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr13_-_9814407 | 0.05 |
ENSMUST00000146059.8
ENSMUST00000110637.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr2_+_112209548 | 0.05 |
ENSMUST00000028552.4
|
Katnbl1
|
katanin p80 subunit B like 1 |
| chr2_+_49341498 | 0.05 |
ENSMUST00000092123.11
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr10_-_43416941 | 0.04 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr14_-_70680882 | 0.04 |
ENSMUST00000000793.13
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr3_-_104419128 | 0.04 |
ENSMUST00000199070.5
ENSMUST00000046316.11 ENSMUST00000198332.2 |
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr2_-_34262012 | 0.04 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr8_+_27513819 | 0.04 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr1_-_168259465 | 0.04 |
ENSMUST00000176540.8
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chr10_+_111309020 | 0.04 |
ENSMUST00000065917.16
ENSMUST00000219961.2 ENSMUST00000217908.2 |
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_93349637 | 0.04 |
ENSMUST00000064257.6
|
Tchh
|
trichohyalin |
| chr9_-_107109108 | 0.04 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr1_+_45350698 | 0.04 |
ENSMUST00000087883.13
|
Col3a1
|
collagen, type III, alpha 1 |
| chr19_-_7360350 | 0.04 |
ENSMUST00000025924.4
|
Spindoc
|
spindlin interactor and repressor of chromatin binding |
| chr6_-_127128007 | 0.04 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr8_-_111754379 | 0.03 |
ENSMUST00000040241.15
|
Ddx19b
|
DEAD box helicase 19b |
| chr3_+_68375495 | 0.03 |
ENSMUST00000182532.8
|
Schip1
|
schwannomin interacting protein 1 |
| chr14_-_49304110 | 0.03 |
ENSMUST00000162175.9
|
Exoc5
|
exocyst complex component 5 |
| chr19_+_40819682 | 0.03 |
ENSMUST00000025983.13
ENSMUST00000119316.2 |
Ccnj
|
cyclin J |
| chr6_-_52194440 | 0.03 |
ENSMUST00000048715.9
|
Hoxa7
|
homeobox A7 |
| chr6_-_144994534 | 0.03 |
ENSMUST00000032402.12
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr2_-_152256947 | 0.03 |
ENSMUST00000099207.5
|
Zcchc3
|
zinc finger, CCHC domain containing 3 |
| chr11_-_106107132 | 0.03 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr12_-_34578842 | 0.03 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr4_+_86792633 | 0.03 |
ENSMUST00000045224.14
ENSMUST00000084433.5 |
Acer2
|
alkaline ceramidase 2 |
| chrX_+_55655111 | 0.03 |
ENSMUST00000144068.8
ENSMUST00000077741.12 ENSMUST00000114784.4 |
Slc9a6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6 |
| chr13_+_45660905 | 0.03 |
ENSMUST00000000260.13
|
Gmpr
|
guanosine monophosphate reductase |
| chr11_-_95896721 | 0.03 |
ENSMUST00000013559.3
|
Igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr17_+_34148485 | 0.03 |
ENSMUST00000047503.16
|
Rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chrX_+_92698469 | 0.03 |
ENSMUST00000113933.9
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr2_+_156038562 | 0.03 |
ENSMUST00000037401.10
|
Phf20
|
PHD finger protein 20 |
| chr17_-_34822649 | 0.03 |
ENSMUST00000015622.8
|
Rnf5
|
ring finger protein 5 |
| chr13_+_35843816 | 0.02 |
ENSMUST00000075220.14
|
Cdyl
|
chromodomain protein, Y chromosome-like |
| chr6_+_14901343 | 0.02 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr6_+_7844759 | 0.02 |
ENSMUST00000040159.6
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr1_+_52158599 | 0.02 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr18_-_81029986 | 0.02 |
ENSMUST00000057950.9
|
Sall3
|
spalt like transcription factor 3 |
| chr6_-_115971914 | 0.02 |
ENSMUST00000015511.15
|
Plxnd1
|
plexin D1 |
| chrX_+_152020744 | 0.02 |
ENSMUST00000112574.9
|
Klf8
|
Kruppel-like factor 8 |
| chrX_-_105055486 | 0.02 |
ENSMUST00000238718.2
ENSMUST00000033583.14 ENSMUST00000151689.9 |
Magt1
|
magnesium transporter 1 |
| chr4_-_82777746 | 0.02 |
ENSMUST00000156055.2
ENSMUST00000030110.15 |
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr1_-_163552693 | 0.02 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr16_-_87229485 | 0.02 |
ENSMUST00000039449.9
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr2_-_52566583 | 0.02 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr7_-_141014477 | 0.02 |
ENSMUST00000106007.10
ENSMUST00000150026.2 ENSMUST00000202840.4 ENSMUST00000133206.9 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr10_+_127575407 | 0.02 |
ENSMUST00000054287.9
|
Zbtb39
|
zinc finger and BTB domain containing 39 |
| chr1_+_20960819 | 0.02 |
ENSMUST00000189400.7
|
Paqr8
|
progestin and adipoQ receptor family member VIII |
| chrX_-_103244784 | 0.02 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr12_-_27392356 | 0.01 |
ENSMUST00000079063.7
|
Sox11
|
SRY (sex determining region Y)-box 11 |
| chr9_-_117081518 | 0.01 |
ENSMUST00000111773.10
ENSMUST00000068962.14 ENSMUST00000044901.14 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr17_-_91400499 | 0.01 |
ENSMUST00000160844.10
|
Nrxn1
|
neurexin I |
| chr12_-_78908409 | 0.01 |
ENSMUST00000021536.9
|
Atp6v1d
|
ATPase, H+ transporting, lysosomal V1 subunit D |
| chrX_-_110606766 | 0.01 |
ENSMUST00000113422.9
ENSMUST00000038472.7 |
Hdx
|
highly divergent homeobox |
| chr3_-_132940647 | 0.01 |
ENSMUST00000147041.10
ENSMUST00000161022.9 |
Arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chrX_+_139565657 | 0.01 |
ENSMUST00000112990.8
ENSMUST00000112988.8 |
Mid2
|
midline 2 |
| chr8_-_111724409 | 0.01 |
ENSMUST00000040416.8
|
Ddx19a
|
DEAD box helicase 19a |
| chr14_-_33169099 | 0.01 |
ENSMUST00000111944.10
ENSMUST00000022504.12 ENSMUST00000111945.9 |
Mapk8
|
mitogen-activated protein kinase 8 |
| chr10_-_123032821 | 0.01 |
ENSMUST00000219619.2
ENSMUST00000020334.9 |
Usp15
|
ubiquitin specific peptidase 15 |
| chr15_-_43034205 | 0.01 |
ENSMUST00000063492.8
ENSMUST00000226810.2 |
Rspo2
|
R-spondin 2 |
| chr8_-_91074971 | 0.01 |
ENSMUST00000109621.10
|
Tox3
|
TOX high mobility group box family member 3 |
| chr8_-_92039850 | 0.01 |
ENSMUST00000047783.14
|
Rpgrip1l
|
Rpgrip1-like |
| chr12_+_117480099 | 0.01 |
ENSMUST00000109691.4
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr15_+_55171138 | 0.01 |
ENSMUST00000023053.12
ENSMUST00000110217.10 |
Col14a1
|
collagen, type XIV, alpha 1 |
| chr11_+_54194624 | 0.01 |
ENSMUST00000093106.12
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr10_-_61219229 | 0.01 |
ENSMUST00000020289.10
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr8_+_24159669 | 0.01 |
ENSMUST00000042352.11
ENSMUST00000207301.2 |
Zmat4
|
zinc finger, matrin type 4 |
| chr9_-_123461593 | 0.01 |
ENSMUST00000026273.11
|
Slc6a20b
|
solute carrier family 6 (neurotransmitter transporter), member 20B |
| chr10_+_58282735 | 0.01 |
ENSMUST00000003310.7
|
Ranbp2
|
RAN binding protein 2 |
| chr4_-_35845204 | 0.01 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr2_+_151923449 | 0.01 |
ENSMUST00000064061.4
|
Scrt2
|
scratch family zinc finger 2 |
| chr1_-_190758321 | 0.00 |
ENSMUST00000191946.2
ENSMUST00000085635.6 |
Flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr19_+_28941292 | 0.00 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr18_+_11052458 | 0.00 |
ENSMUST00000047762.10
|
Gata6
|
GATA binding protein 6 |
| chr16_-_4950285 | 0.00 |
ENSMUST00000035672.5
|
Ppl
|
periplakin |
| chr7_+_15864265 | 0.00 |
ENSMUST00000168693.3
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr17_-_81035453 | 0.00 |
ENSMUST00000234133.2
ENSMUST00000112389.9 ENSMUST00000025089.9 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chrX_-_13712746 | 0.00 |
ENSMUST00000115436.9
ENSMUST00000033321.11 ENSMUST00000115438.10 |
Cask
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr12_-_104718159 | 0.00 |
ENSMUST00000041987.7
|
Dicer1
|
dicer 1, ribonuclease type III |
| chrX_-_160942713 | 0.00 |
ENSMUST00000087085.10
|
Nhs
|
NHS actin remodeling regulator |
| chr3_-_91990439 | 0.00 |
ENSMUST00000058150.8
|
Lor
|
loricrin |
| chr5_+_13449276 | 0.00 |
ENSMUST00000030714.9
ENSMUST00000141968.2 |
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_+_137770813 | 0.00 |
ENSMUST00000163080.3
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr6_-_120470768 | 0.00 |
ENSMUST00000178687.2
|
Tmem121b
|
transmembrane protein 121B |
| chr16_-_97412169 | 0.00 |
ENSMUST00000232141.2
ENSMUST00000000395.8 |
Tmprss2
|
transmembrane protease, serine 2 |
| chr6_+_4505493 | 0.00 |
ENSMUST00000031668.10
|
Col1a2
|
collagen, type I, alpha 2 |
| chr6_+_4747298 | 0.00 |
ENSMUST00000166678.2
ENSMUST00000176204.8 |
Peg10
|
paternally expressed 10 |
| chrX_-_7054952 | 0.00 |
ENSMUST00000004428.14
|
Clcn5
|
chloride channel, voltage-sensitive 5 |
| chr15_+_87509413 | 0.00 |
ENSMUST00000068088.8
|
Tafa5
|
TAFA chemokine like family member 5 |
| chr14_-_124914516 | 0.00 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr11_-_57723502 | 0.00 |
ENSMUST00000160392.9
ENSMUST00000108845.3 |
Hand1
|
heart and neural crest derivatives expressed 1 |
| chr10_+_79518002 | 0.00 |
ENSMUST00000020550.13
|
Cdc34
|
cell division cycle 34 |
| chr8_+_14145848 | 0.00 |
ENSMUST00000152652.8
ENSMUST00000133298.8 |
Dlgap2
|
DLG associated protein 2 |
| chr7_+_90075762 | 0.00 |
ENSMUST00000061391.9
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr16_-_67417768 | 0.00 |
ENSMUST00000114292.8
ENSMUST00000120898.8 |
Cadm2
|
cell adhesion molecule 2 |
| chr10_-_23225781 | 0.00 |
ENSMUST00000092665.12
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr4_-_139560831 | 0.00 |
ENSMUST00000030508.14
|
Pax7
|
paired box 7 |
| chr4_+_21931290 | 0.00 |
ENSMUST00000029908.8
|
Faxc
|
failed axon connections homolog |
| chr3_+_144998233 | 0.00 |
ENSMUST00000029848.5
ENSMUST00000139001.2 |
Col24a1
|
collagen, type XXIV, alpha 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AGGUAGU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0060574 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.0 | 0.1 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |