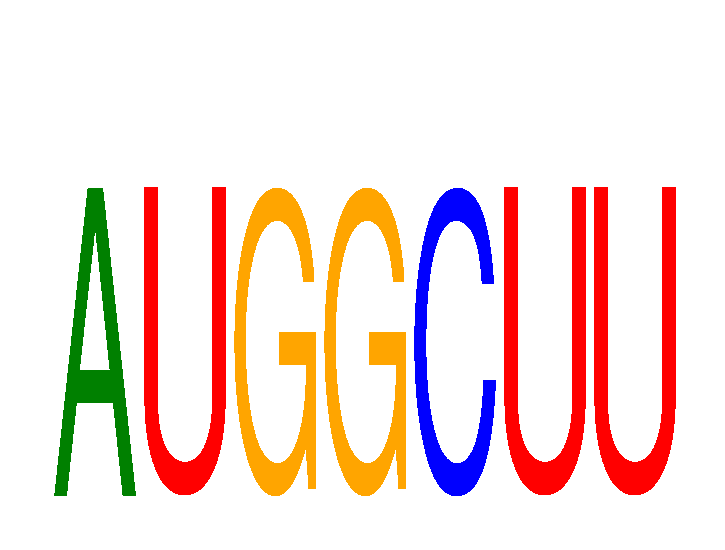Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for AUGGCUU
Z-value: 0.66
miRNA associated with seed AUGGCUU
| Name | miRBASE accession |
|---|---|
|
mmu-miR-135a-5p
|
MIMAT0000147 |
|
mmu-miR-135b-5p
|
MIMAT0000612 |
Activity profile of AUGGCUU motif
Sorted Z-values of AUGGCUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_48358267 | 0.36 |
ENSMUST00000073150.6
|
Peli2
|
pellino 2 |
| chr13_-_40887244 | 0.29 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr7_+_43959637 | 0.28 |
ENSMUST00000107938.8
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr13_+_54519161 | 0.28 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr11_+_68979332 | 0.28 |
ENSMUST00000117780.2
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr16_+_37909363 | 0.28 |
ENSMUST00000023507.13
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr7_+_27878894 | 0.24 |
ENSMUST00000085901.13
ENSMUST00000172761.8 |
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr15_-_58953838 | 0.24 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr6_-_39183712 | 0.23 |
ENSMUST00000002305.9
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chrX_+_7688528 | 0.22 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr1_-_133728779 | 0.21 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr11_+_98828495 | 0.20 |
ENSMUST00000107475.9
ENSMUST00000068133.10 |
Rara
|
retinoic acid receptor, alpha |
| chr8_+_36054919 | 0.18 |
ENSMUST00000037666.6
|
Mfhas1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr7_+_121666388 | 0.18 |
ENSMUST00000033158.6
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr1_+_166828982 | 0.17 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr9_+_43655230 | 0.16 |
ENSMUST00000034510.9
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr1_-_162567919 | 0.16 |
ENSMUST00000182331.2
ENSMUST00000183011.8 ENSMUST00000182593.8 ENSMUST00000182149.8 |
Prrc2c
|
proline-rich coiled-coil 2C |
| chr6_+_117145496 | 0.16 |
ENSMUST00000112866.8
ENSMUST00000112871.8 ENSMUST00000073043.5 |
Cxcl12
|
chemokine (C-X-C motif) ligand 12 |
| chr14_-_30075424 | 0.16 |
ENSMUST00000224198.3
ENSMUST00000238675.2 ENSMUST00000112249.10 ENSMUST00000224785.3 |
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr9_+_108685555 | 0.16 |
ENSMUST00000035218.9
ENSMUST00000195323.2 ENSMUST00000194819.2 |
Nckipsd
|
NCK interacting protein with SH3 domain |
| chr3_-_119576911 | 0.15 |
ENSMUST00000197464.5
ENSMUST00000198403.2 ENSMUST00000029780.12 |
Ptbp2
|
polypyrimidine tract binding protein 2 |
| chr11_+_44508137 | 0.14 |
ENSMUST00000109268.2
ENSMUST00000101326.10 ENSMUST00000081265.12 |
Ebf1
|
early B cell factor 1 |
| chrX_+_72716756 | 0.14 |
ENSMUST00000033752.14
ENSMUST00000114467.9 |
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr9_+_59658156 | 0.14 |
ENSMUST00000136740.8
ENSMUST00000135298.8 ENSMUST00000128341.2 |
Myo9a
|
myosin IXa |
| chr9_+_44410417 | 0.14 |
ENSMUST00000074989.7
ENSMUST00000218913.2 |
Bcl9l
|
B cell CLL/lymphoma 9-like |
| chr17_+_15163446 | 0.13 |
ENSMUST00000052691.9
|
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene |
| chr15_+_85942928 | 0.13 |
ENSMUST00000138134.8
|
Gramd4
|
GRAM domain containing 4 |
| chr16_+_51851588 | 0.13 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr4_+_125384481 | 0.13 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr9_+_109760528 | 0.12 |
ENSMUST00000035055.15
|
Map4
|
microtubule-associated protein 4 |
| chr5_+_130477642 | 0.12 |
ENSMUST00000111288.4
|
Caln1
|
calneuron 1 |
| chr9_+_53448322 | 0.12 |
ENSMUST00000035850.8
|
Npat
|
nuclear protein in the AT region |
| chr8_-_84963653 | 0.12 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr16_+_32882882 | 0.11 |
ENSMUST00000023497.3
|
Lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr11_-_86248395 | 0.11 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13 |
| chr6_-_83418656 | 0.11 |
ENSMUST00000089622.11
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr10_+_56253418 | 0.11 |
ENSMUST00000068581.9
ENSMUST00000217789.2 |
Gja1
|
gap junction protein, alpha 1 |
| chr4_-_126323291 | 0.11 |
ENSMUST00000069097.13
|
Ago3
|
argonaute RISC catalytic subunit 3 |
| chr4_-_141450710 | 0.10 |
ENSMUST00000102484.5
ENSMUST00000177592.2 |
Ddi2
|
DNA-damage inducible protein 2 |
| chr1_-_125839897 | 0.10 |
ENSMUST00000159417.2
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr4_-_41569500 | 0.10 |
ENSMUST00000108049.9
ENSMUST00000108052.10 ENSMUST00000108050.2 |
Fam219a
|
family with sequence similarity 219, member A |
| chr15_+_12205095 | 0.10 |
ENSMUST00000038172.16
|
Mtmr12
|
myotubularin related protein 12 |
| chr14_+_47514368 | 0.09 |
ENSMUST00000065562.6
|
Socs4
|
suppressor of cytokine signaling 4 |
| chr13_-_3968157 | 0.09 |
ENSMUST00000223258.2
ENSMUST00000091853.12 |
Net1
|
neuroepithelial cell transforming gene 1 |
| chr3_+_106455114 | 0.09 |
ENSMUST00000067630.13
ENSMUST00000134396.8 ENSMUST00000121034.8 ENSMUST00000029507.13 ENSMUST00000144746.8 ENSMUST00000132923.8 ENSMUST00000151465.2 |
Dram2
|
DNA-damage regulated autophagy modulator 2 |
| chr3_-_19365431 | 0.09 |
ENSMUST00000099195.10
|
Pde7a
|
phosphodiesterase 7A |
| chr3_-_133250129 | 0.09 |
ENSMUST00000196398.5
ENSMUST00000098603.8 |
Tet2
|
tet methylcytosine dioxygenase 2 |
| chr5_+_64960705 | 0.09 |
ENSMUST00000165536.8
|
Klf3
|
Kruppel-like factor 3 (basic) |
| chr11_+_95603494 | 0.09 |
ENSMUST00000107717.8
|
Zfp652
|
zinc finger protein 652 |
| chr9_+_102885156 | 0.09 |
ENSMUST00000035148.13
|
Slco2a1
|
solute carrier organic anion transporter family, member 2a1 |
| chr2_-_167191087 | 0.09 |
ENSMUST00000109221.9
|
B4galt5
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5 |
| chr18_-_15196612 | 0.09 |
ENSMUST00000168989.9
|
Kctd1
|
potassium channel tetramerisation domain containing 1 |
| chr11_-_78056347 | 0.09 |
ENSMUST00000017530.4
|
Traf4
|
TNF receptor associated factor 4 |
| chr9_-_95632387 | 0.09 |
ENSMUST00000189137.7
ENSMUST00000053785.10 |
Trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr7_+_24967094 | 0.09 |
ENSMUST00000169266.8
|
Cic
|
capicua transcriptional repressor |
| chr2_-_90900628 | 0.09 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr4_-_141412910 | 0.09 |
ENSMUST00000105782.2
|
Rsc1a1
|
regulatory solute carrier protein, family 1, member 1 |
| chr7_+_4743114 | 0.08 |
ENSMUST00000098853.9
ENSMUST00000130215.8 ENSMUST00000108582.10 ENSMUST00000108583.9 |
Kmt5c
|
lysine methyltransferase 5C |
| chr8_-_106553822 | 0.08 |
ENSMUST00000239468.2
ENSMUST00000041400.6 |
Ranbp10
|
RAN binding protein 10 |
| chr1_+_131935342 | 0.08 |
ENSMUST00000086556.12
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr4_-_34882917 | 0.08 |
ENSMUST00000098163.9
ENSMUST00000047950.6 |
Zfp292
|
zinc finger protein 292 |
| chr11_-_69692542 | 0.08 |
ENSMUST00000011285.11
ENSMUST00000102585.2 |
Fgf11
|
fibroblast growth factor 11 |
| chr8_-_125390107 | 0.08 |
ENSMUST00000034464.8
|
2310022B05Rik
|
RIKEN cDNA 2310022B05 gene |
| chr9_+_21077010 | 0.08 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr1_-_152642032 | 0.08 |
ENSMUST00000111859.8
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr8_-_17585263 | 0.08 |
ENSMUST00000082104.7
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr9_+_45029080 | 0.08 |
ENSMUST00000170998.9
ENSMUST00000093855.4 |
Scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr2_-_33321306 | 0.08 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr1_-_105284383 | 0.08 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr14_-_57983511 | 0.08 |
ENSMUST00000173990.8
ENSMUST00000022531.14 |
Lats2
|
large tumor suppressor 2 |
| chr1_-_156767123 | 0.08 |
ENSMUST00000189316.7
ENSMUST00000190648.7 ENSMUST00000172057.8 ENSMUST00000191605.7 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr18_+_7869066 | 0.08 |
ENSMUST00000171486.8
ENSMUST00000170932.8 ENSMUST00000167020.8 |
Wac
|
WW domain containing adaptor with coiled-coil |
| chr3_+_129326004 | 0.07 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr10_-_80413119 | 0.07 |
ENSMUST00000038558.9
|
Klf16
|
Kruppel-like factor 16 |
| chr1_-_13442658 | 0.07 |
ENSMUST00000081713.11
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr1_+_59802543 | 0.07 |
ENSMUST00000087435.7
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chrX_-_20816841 | 0.07 |
ENSMUST00000009550.14
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr7_+_28092370 | 0.07 |
ENSMUST00000003529.9
|
Paf1
|
Paf1, RNA polymerase II complex component |
| chr15_-_28025920 | 0.07 |
ENSMUST00000090247.7
|
Trio
|
triple functional domain (PTPRF interacting) |
| chr5_-_135963408 | 0.07 |
ENSMUST00000198270.2
ENSMUST00000055808.6 |
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
| chr8_+_107662352 | 0.07 |
ENSMUST00000212524.2
ENSMUST00000047425.5 |
Sntb2
|
syntrophin, basic 2 |
| chr8_+_85763534 | 0.07 |
ENSMUST00000093360.12
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr16_+_17051423 | 0.07 |
ENSMUST00000090190.14
ENSMUST00000232082.2 ENSMUST00000232426.2 |
Hic2
Gm49573
|
hypermethylated in cancer 2 predicted gene, 49573 |
| chr8_-_36716445 | 0.07 |
ENSMUST00000239119.2
ENSMUST00000065297.6 |
Lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr8_-_71229293 | 0.07 |
ENSMUST00000034296.15
|
Pik3r2
|
phosphoinositide-3-kinase regulatory subunit 2 |
| chr4_+_83443777 | 0.07 |
ENSMUST00000053414.13
ENSMUST00000231339.2 ENSMUST00000126429.8 |
Ccdc171
|
coiled-coil domain containing 171 |
| chr5_-_21156766 | 0.07 |
ENSMUST00000036489.10
|
Rsbn1l
|
round spermatid basic protein 1-like |
| chr10_+_83379805 | 0.07 |
ENSMUST00000038388.7
|
Washc4
|
WASH complex subunit 4 |
| chr9_+_8544143 | 0.07 |
ENSMUST00000050433.8
ENSMUST00000217462.2 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr1_+_34840785 | 0.06 |
ENSMUST00000047664.16
ENSMUST00000211073.2 |
Arhgef4
SMIM39
|
Rho guanine nucleotide exchange factor (GEF) 4 novel protein |
| chr2_+_37406630 | 0.06 |
ENSMUST00000065441.7
|
Gpr21
|
G protein-coupled receptor 21 |
| chr17_-_88105422 | 0.06 |
ENSMUST00000055221.9
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr2_-_179618439 | 0.06 |
ENSMUST00000041618.13
ENSMUST00000227325.2 |
Taf4
|
TATA-box binding protein associated factor 4 |
| chr9_-_106035308 | 0.06 |
ENSMUST00000159809.2
ENSMUST00000162562.2 ENSMUST00000036382.13 |
Glyctk
|
glycerate kinase |
| chr5_+_138083345 | 0.06 |
ENSMUST00000019660.11
ENSMUST00000066617.12 ENSMUST00000110963.8 |
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr1_+_127701901 | 0.06 |
ENSMUST00000112570.2
ENSMUST00000027587.15 |
Ccnt2
|
cyclin T2 |
| chr11_+_67477347 | 0.06 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr2_-_167334746 | 0.06 |
ENSMUST00000109211.9
ENSMUST00000057627.16 |
Spata2
|
spermatogenesis associated 2 |
| chr16_-_97723753 | 0.06 |
ENSMUST00000170757.3
|
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr16_-_13485986 | 0.06 |
ENSMUST00000058884.10
|
Parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr12_+_16944896 | 0.06 |
ENSMUST00000020904.8
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2 |
| chr8_+_11808349 | 0.06 |
ENSMUST00000074856.13
ENSMUST00000098938.9 |
Arhgef7
|
Rho guanine nucleotide exchange factor (GEF7) |
| chr4_-_55532453 | 0.06 |
ENSMUST00000132746.2
ENSMUST00000107619.3 |
Klf4
|
Kruppel-like factor 4 (gut) |
| chr2_+_174485327 | 0.06 |
ENSMUST00000059452.6
|
Zfp831
|
zinc finger protein 831 |
| chr7_+_18725170 | 0.06 |
ENSMUST00000059331.9
ENSMUST00000131087.2 |
Mypop
|
Myb-related transcription factor, partner of profilin |
| chr19_+_29499671 | 0.06 |
ENSMUST00000043610.13
|
Ric1
|
RAB6A GEF complex partner 1 |
| chr19_+_10666085 | 0.06 |
ENSMUST00000237240.2
ENSMUST00000235927.2 ENSMUST00000087951.7 ENSMUST00000237437.2 ENSMUST00000235921.2 |
Vps37c
|
vacuolar protein sorting 37C |
| chr14_-_68362284 | 0.06 |
ENSMUST00000111089.8
ENSMUST00000022638.6 |
Nefm
|
neurofilament, medium polypeptide |
| chr5_-_143166476 | 0.06 |
ENSMUST00000049861.11
ENSMUST00000165318.4 |
Rbak
|
RB-associated KRAB zinc finger |
| chr1_+_37338964 | 0.06 |
ENSMUST00000027287.11
ENSMUST00000140264.8 |
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I |
| chr5_-_124465946 | 0.05 |
ENSMUST00000031344.13
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr9_+_30941924 | 0.05 |
ENSMUST00000216649.2
ENSMUST00000115222.10 |
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr6_-_136150076 | 0.05 |
ENSMUST00000053880.13
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chr2_-_9883391 | 0.05 |
ENSMUST00000102976.4
|
Gata3
|
GATA binding protein 3 |
| chr7_-_46445305 | 0.05 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr6_+_72324281 | 0.05 |
ENSMUST00000077783.5
|
0610030E20Rik
|
RIKEN cDNA 0610030E20 gene |
| chr18_-_10182007 | 0.05 |
ENSMUST00000067947.7
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr3_+_96465265 | 0.05 |
ENSMUST00000074519.13
ENSMUST00000049093.8 |
Txnip
|
thioredoxin interacting protein |
| chr8_-_61436249 | 0.05 |
ENSMUST00000004430.14
ENSMUST00000110301.2 ENSMUST00000093490.9 |
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr10_+_126814542 | 0.05 |
ENSMUST00000105256.10
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr15_-_103248512 | 0.05 |
ENSMUST00000168828.3
|
Zfp385a
|
zinc finger protein 385A |
| chr8_-_110464345 | 0.05 |
ENSMUST00000212605.2
ENSMUST00000093162.4 ENSMUST00000212726.2 |
Atxn1l
|
ataxin 1-like |
| chr4_-_135749032 | 0.05 |
ENSMUST00000030427.6
|
Eloa
|
elongin A |
| chr15_+_78761360 | 0.05 |
ENSMUST00000041587.8
|
Gga1
|
golgi associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr6_-_35516700 | 0.05 |
ENSMUST00000201026.2
ENSMUST00000031866.9 |
Mtpn
|
myotrophin |
| chr2_-_154249997 | 0.05 |
ENSMUST00000028991.7
ENSMUST00000109728.8 |
Snta1
|
syntrophin, acidic 1 |
| chr5_+_88868714 | 0.05 |
ENSMUST00000113229.8
ENSMUST00000006424.8 |
Mob1b
|
MOB kinase activator 1B |
| chr10_-_127047396 | 0.05 |
ENSMUST00000013970.9
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr16_-_58343789 | 0.05 |
ENSMUST00000137035.8
ENSMUST00000149456.8 |
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr15_+_10177709 | 0.05 |
ENSMUST00000124470.8
|
Prlr
|
prolactin receptor |
| chr2_-_181101158 | 0.05 |
ENSMUST00000155535.2
ENSMUST00000029106.13 ENSMUST00000087409.10 |
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr2_-_126718129 | 0.05 |
ENSMUST00000103224.10
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr14_+_21549798 | 0.05 |
ENSMUST00000182855.8
ENSMUST00000182405.9 ENSMUST00000069648.14 |
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr4_-_35157405 | 0.04 |
ENSMUST00000102975.10
|
Mob3b
|
MOB kinase activator 3B |
| chr10_-_127124718 | 0.04 |
ENSMUST00000156208.2
ENSMUST00000026476.13 |
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr10_-_121422635 | 0.04 |
ENSMUST00000219493.2
ENSMUST00000020316.4 |
Tbk1
|
TANK-binding kinase 1 |
| chr3_+_153679073 | 0.04 |
ENSMUST00000089948.6
|
Slc44a5
|
solute carrier family 44, member 5 |
| chr11_-_94212783 | 0.04 |
ENSMUST00000166312.8
ENSMUST00000107821.9 ENSMUST00000021226.14 ENSMUST00000107820.2 |
Luc7l3
|
LUC7-like 3 (S. cerevisiae) |
| chr11_-_5331734 | 0.04 |
ENSMUST00000172492.8
|
Znrf3
|
zinc and ring finger 3 |
| chr19_+_29229147 | 0.04 |
ENSMUST00000025705.7
ENSMUST00000065796.10 ENSMUST00000236990.2 |
Jak2
|
Janus kinase 2 |
| chr18_-_61310066 | 0.04 |
ENSMUST00000235480.2
ENSMUST00000091884.6 |
Hmgxb3
|
HMG box domain containing 3 |
| chr4_+_139107911 | 0.04 |
ENSMUST00000165860.8
ENSMUST00000097822.10 |
Ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr6_-_38230890 | 0.04 |
ENSMUST00000117556.8
ENSMUST00000169256.5 |
D630045J12Rik
|
RIKEN cDNA D630045J12 gene |
| chr9_-_36637670 | 0.04 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr4_-_70453140 | 0.04 |
ENSMUST00000107359.9
|
Megf9
|
multiple EGF-like-domains 9 |
| chr8_+_106002772 | 0.04 |
ENSMUST00000014920.8
|
Nol3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr9_+_100956734 | 0.04 |
ENSMUST00000085177.5
|
Msl2
|
MSL complex subunit 2 |
| chr19_+_55730696 | 0.04 |
ENSMUST00000153888.9
ENSMUST00000127233.9 ENSMUST00000061496.17 ENSMUST00000111656.8 ENSMUST00000111657.11 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr11_-_86435579 | 0.03 |
ENSMUST00000138810.3
ENSMUST00000058286.9 ENSMUST00000154617.8 |
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1 |
| chr7_-_121306476 | 0.03 |
ENSMUST00000046929.7
|
Usp31
|
ubiquitin specific peptidase 31 |
| chr2_-_79959178 | 0.03 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr4_-_34687381 | 0.03 |
ENSMUST00000029970.14
|
Slc35a1
|
solute carrier family 35 (CMP-sialic acid transporter), member 1 |
| chr9_+_121946321 | 0.03 |
ENSMUST00000119215.9
ENSMUST00000146832.8 ENSMUST00000118886.9 ENSMUST00000120173.9 ENSMUST00000139181.2 |
Snrk
|
SNF related kinase |
| chr15_-_73056713 | 0.03 |
ENSMUST00000044113.12
|
Ago2
|
argonaute RISC catalytic subunit 2 |
| chr9_-_42035560 | 0.03 |
ENSMUST00000060989.9
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr1_-_173195236 | 0.03 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr11_+_95715295 | 0.03 |
ENSMUST00000150134.2
ENSMUST00000054173.4 |
Phospho1
|
phosphatase, orphan 1 |
| chr14_+_69574623 | 0.03 |
ENSMUST00000184973.8
ENSMUST00000184314.8 |
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr15_-_64184485 | 0.03 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr2_+_119803180 | 0.03 |
ENSMUST00000066058.8
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr7_-_67022520 | 0.03 |
ENSMUST00000156690.8
ENSMUST00000107476.8 ENSMUST00000076325.12 ENSMUST00000032776.15 ENSMUST00000133074.2 |
Mef2a
|
myocyte enhancer factor 2A |
| chr15_-_53209513 | 0.03 |
ENSMUST00000077273.9
|
Ext1
|
exostosin glycosyltransferase 1 |
| chr6_-_38276157 | 0.03 |
ENSMUST00000058524.3
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like |
| chr9_+_65816206 | 0.03 |
ENSMUST00000205379.2
ENSMUST00000206048.2 ENSMUST00000034949.10 ENSMUST00000154589.2 |
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr11_+_94881861 | 0.03 |
ENSMUST00000038696.12
|
Ppp1r9b
|
protein phosphatase 1, regulatory subunit 9B |
| chrX_+_40490005 | 0.03 |
ENSMUST00000115103.9
ENSMUST00000076349.12 |
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr19_+_44919585 | 0.03 |
ENSMUST00000096053.5
|
Slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr2_+_32178255 | 0.03 |
ENSMUST00000239447.2
ENSMUST00000239474.2 ENSMUST00000113377.8 |
Golga2
|
golgi autoantigen, golgin subfamily a, 2 |
| chr15_-_78004211 | 0.03 |
ENSMUST00000019290.3
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2 |
| chr11_+_21041291 | 0.03 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr14_+_120715855 | 0.03 |
ENSMUST00000062117.14
|
Rap2a
|
RAS related protein 2a |
| chr7_+_24181416 | 0.03 |
ENSMUST00000068023.8
|
Cadm4
|
cell adhesion molecule 4 |
| chr4_+_57845240 | 0.03 |
ENSMUST00000102903.8
ENSMUST00000107598.9 |
Pakap
|
paralemmin A kinase anchor protein |
| chr14_+_69792873 | 0.03 |
ENSMUST00000185072.8
ENSMUST00000064846.8 |
Entpd4b
|
ectonucleoside triphosphate diphosphohydrolase 4B |
| chr3_+_122039206 | 0.03 |
ENSMUST00000029769.14
|
Gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr1_+_36107452 | 0.03 |
ENSMUST00000088174.4
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr4_+_100926170 | 0.03 |
ENSMUST00000106955.2
ENSMUST00000038463.15 |
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr13_-_89890609 | 0.03 |
ENSMUST00000109546.9
|
Vcan
|
versican |
| chr18_+_21134302 | 0.03 |
ENSMUST00000234107.2
ENSMUST00000072847.12 |
Rnf138
|
ring finger protein 138 |
| chr16_+_87495792 | 0.02 |
ENSMUST00000026703.6
|
Bach1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr16_+_49675969 | 0.02 |
ENSMUST00000229101.2
ENSMUST00000230836.2 ENSMUST00000229206.2 ENSMUST00000084838.14 ENSMUST00000230281.2 |
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr5_-_136596299 | 0.02 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1 |
| chr10_-_86334700 | 0.02 |
ENSMUST00000120638.8
|
Syn3
|
synapsin III |
| chr10_-_128237087 | 0.02 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr2_+_172187485 | 0.02 |
ENSMUST00000028995.5
|
Fam210b
|
family with sequence similarity 210, member B |
| chr19_-_3625698 | 0.02 |
ENSMUST00000172362.3
ENSMUST00000025846.16 ENSMUST00000226109.2 ENSMUST00000113997.9 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr4_+_148888877 | 0.02 |
ENSMUST00000094464.10
ENSMUST00000122222.8 |
Casz1
|
castor zinc finger 1 |
| chr10_-_121146940 | 0.02 |
ENSMUST00000064107.7
|
Tbc1d30
|
TBC1 domain family, member 30 |
| chr19_-_6899173 | 0.02 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr7_-_141649003 | 0.02 |
ENSMUST00000039926.10
|
Dusp8
|
dual specificity phosphatase 8 |
| chrX_+_167819606 | 0.02 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chrX_+_72546680 | 0.02 |
ENSMUST00000033744.12
ENSMUST00000088429.8 ENSMUST00000114479.2 |
Atp2b3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr2_-_169973076 | 0.02 |
ENSMUST00000063710.13
|
Zfp217
|
zinc finger protein 217 |
| chr4_+_57637817 | 0.02 |
ENSMUST00000150412.4
|
Pakap
|
paralemmin A kinase anchor protein |
| chr14_-_60324265 | 0.02 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr9_-_70048766 | 0.02 |
ENSMUST00000034749.16
|
Fam81a
|
family with sequence similarity 81, member A |
| chr9_+_96001415 | 0.02 |
ENSMUST00000085217.12
ENSMUST00000122383.3 |
Gk5
|
glycerol kinase 5 (putative) |
| chr4_-_155306992 | 0.02 |
ENSMUST00000084103.10
ENSMUST00000030917.6 |
Ski
|
ski sarcoma viral oncogene homolog (avian) |
| chr2_+_129642371 | 0.02 |
ENSMUST00000165413.9
ENSMUST00000166282.3 |
Stk35
|
serine/threonine kinase 35 |
| chr11_+_101066867 | 0.02 |
ENSMUST00000103109.4
|
Cntnap1
|
contactin associated protein-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AUGGCUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.2 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:0015881 | creatine transport(GO:0015881) |
| 0.0 | 0.2 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.3 | GO:0002278 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.3 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.0 | GO:0071317 | cellular response to isoquinoline alkaloid(GO:0071317) |
| 0.0 | 0.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.0 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.0 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.0 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.2 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0034066 | RIC1-RGP1 guanyl-nucleotide exchange factor complex(GO:0034066) |
| 0.0 | 0.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.2 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |