Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Aire
Z-value: 0.64
Transcription factors associated with Aire
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Aire
|
ENSMUSG00000000731.16 | autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Aire | mm39_v1_chr10_-_77879414_77879445 | -0.05 | 9.3e-01 | Click! |
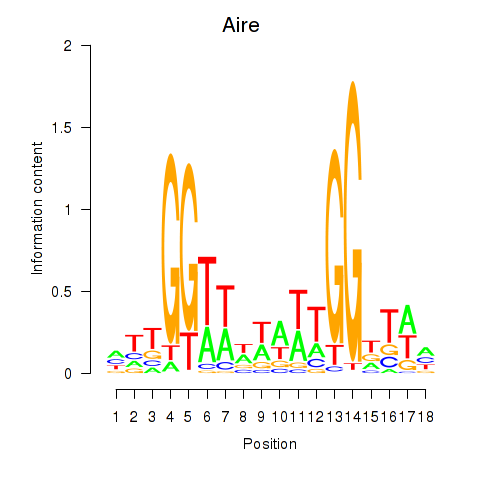
Activity profile of Aire motif
Sorted Z-values of Aire motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_21934675 | 0.95 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chrM_+_11735 | 0.67 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrM_+_14138 | 0.54 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chrM_-_14061 | 0.53 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr16_-_36228798 | 0.41 |
ENSMUST00000023619.8
|
Stfa2
|
stefin A2 |
| chr7_+_142014546 | 0.36 |
ENSMUST00000105968.8
ENSMUST00000018963.11 ENSMUST00000105967.8 |
Lsp1
|
lymphocyte specific 1 |
| chr9_-_110818679 | 0.36 |
ENSMUST00000084922.6
ENSMUST00000199891.2 |
Rtp3
|
receptor transporter protein 3 |
| chrM_+_7006 | 0.33 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr9_+_45924120 | 0.31 |
ENSMUST00000120463.9
ENSMUST00000120247.8 |
Sik3
|
SIK family kinase 3 |
| chr2_-_88937158 | 0.29 |
ENSMUST00000099806.3
ENSMUST00000213288.2 |
Olfr1220
|
olfactory receptor 1220 |
| chr7_-_102774821 | 0.29 |
ENSMUST00000211075.3
ENSMUST00000215304.2 ENSMUST00000213281.2 |
Olfr586
|
olfactory receptor 586 |
| chr9_+_45924105 | 0.29 |
ENSMUST00000126865.8
|
Sik3
|
SIK family kinase 3 |
| chr12_+_37292029 | 0.25 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr9_-_49479791 | 0.24 |
ENSMUST00000194252.6
|
Ncam1
|
neural cell adhesion molecule 1 |
| chr13_+_49658249 | 0.24 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr11_+_69805005 | 0.23 |
ENSMUST00000057884.6
|
Gps2
|
G protein pathway suppressor 2 |
| chr19_-_5713701 | 0.23 |
ENSMUST00000164304.9
ENSMUST00000237544.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr16_-_36695166 | 0.22 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2 |
| chr5_-_84565218 | 0.22 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr8_+_65399831 | 0.22 |
ENSMUST00000026595.13
ENSMUST00000209852.2 ENSMUST00000079896.9 |
Tmem192
|
transmembrane protein 192 |
| chr11_+_62349238 | 0.21 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr11_-_69586884 | 0.21 |
ENSMUST00000180587.8
|
Tnfsfm13
|
tumor necrosis factor (ligand) superfamily, membrane-bound member 13 |
| chrX_-_37665041 | 0.20 |
ENSMUST00000050083.6
ENSMUST00000115118.8 |
Cul4b
|
cullin 4B |
| chr10_+_3822667 | 0.19 |
ENSMUST00000136671.8
ENSMUST00000042438.13 |
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr15_+_89095724 | 0.18 |
ENSMUST00000227951.2
ENSMUST00000226221.2 ENSMUST00000238818.2 ENSMUST00000228284.2 |
Ppp6r2
|
protein phosphatase 6, regulatory subunit 2 |
| chr17_-_59320257 | 0.18 |
ENSMUST00000174122.2
ENSMUST00000025065.12 |
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr7_+_28869770 | 0.18 |
ENSMUST00000033886.8
ENSMUST00000209019.2 ENSMUST00000208330.2 |
Ggn
|
gametogenetin |
| chr1_-_5140504 | 0.17 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr7_-_126275529 | 0.17 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr2_+_89804937 | 0.16 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr7_-_102507962 | 0.15 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr9_-_19452372 | 0.14 |
ENSMUST00000213834.2
|
Olfr853
|
olfactory receptor 853 |
| chr16_-_18904240 | 0.14 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr9_+_108883907 | 0.14 |
ENSMUST00000154184.5
|
Shisa5
|
shisa family member 5 |
| chr6_+_11907808 | 0.14 |
ENSMUST00000155037.4
|
Phf14
|
PHD finger protein 14 |
| chr11_+_105866030 | 0.14 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr17_+_18518361 | 0.14 |
ENSMUST00000231938.2
ENSMUST00000079206.8 ENSMUST00000231879.3 |
Vmn2r93
|
vomeronasal 2, receptor 93 |
| chr7_-_19398930 | 0.13 |
ENSMUST00000055242.11
|
Clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr2_+_172090412 | 0.12 |
ENSMUST00000038532.2
|
Mc3r
|
melanocortin 3 receptor |
| chr2_+_164790139 | 0.11 |
ENSMUST00000017881.3
|
Mmp9
|
matrix metallopeptidase 9 |
| chr5_-_100521343 | 0.11 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr17_+_21869545 | 0.11 |
ENSMUST00000232827.2
|
Zfp983
|
zinc finger protein 983 |
| chr9_-_108140925 | 0.11 |
ENSMUST00000171412.7
ENSMUST00000195429.6 ENSMUST00000080435.9 |
Dag1
|
dystroglycan 1 |
| chr16_-_91415873 | 0.10 |
ENSMUST00000143058.2
ENSMUST00000049244.10 ENSMUST00000169982.2 ENSMUST00000133731.2 |
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr11_-_49077864 | 0.10 |
ENSMUST00000153999.3
ENSMUST00000066531.13 |
Btnl9
|
butyrophilin-like 9 |
| chr11_+_97206542 | 0.10 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr10_-_127587576 | 0.10 |
ENSMUST00000079692.6
|
Gpr182
|
G protein-coupled receptor 182 |
| chr19_-_5713728 | 0.09 |
ENSMUST00000169854.2
|
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr17_+_35455532 | 0.09 |
ENSMUST00000068261.9
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2 |
| chr10_-_111833138 | 0.09 |
ENSMUST00000074805.12
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr4_-_46566431 | 0.08 |
ENSMUST00000030021.14
ENSMUST00000107757.8 |
Coro2a
|
coronin, actin binding protein 2A |
| chr13_+_64309675 | 0.08 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr2_-_134485936 | 0.08 |
ENSMUST00000110120.2
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr1_-_85247864 | 0.08 |
ENSMUST00000159582.8
ENSMUST00000161267.8 |
C130026I21Rik
|
RIKEN cDNA C130026I21 gene |
| chr3_+_107008867 | 0.08 |
ENSMUST00000038695.6
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr11_-_96002784 | 0.08 |
ENSMUST00000097162.6
ENSMUST00000068686.13 |
Calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr2_+_87854404 | 0.07 |
ENSMUST00000217006.2
|
Olfr1161
|
olfactory receptor 1161 |
| chr14_+_21531494 | 0.07 |
ENSMUST00000182996.2
|
Kat6b
|
K(lysine) acetyltransferase 6B |
| chr2_-_134485999 | 0.07 |
ENSMUST00000110119.2
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr17_+_57412325 | 0.06 |
ENSMUST00000039490.9
|
Tnfsf9
|
tumor necrosis factor (ligand) superfamily, member 9 |
| chr6_+_135339929 | 0.06 |
ENSMUST00000032330.16
|
Emp1
|
epithelial membrane protein 1 |
| chr10_+_129263175 | 0.06 |
ENSMUST00000204529.3
|
Olfr786
|
olfactory receptor 786 |
| chr2_-_150531280 | 0.06 |
ENSMUST00000046095.10
|
Vsx1
|
visual system homeobox 1 |
| chr11_-_83177548 | 0.06 |
ENSMUST00000163961.3
|
Slfn14
|
schlafen 14 |
| chr2_-_93841152 | 0.06 |
ENSMUST00000111240.8
|
Alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr19_-_5713648 | 0.06 |
ENSMUST00000080824.13
ENSMUST00000237874.2 ENSMUST00000071857.13 ENSMUST00000236464.2 |
Sipa1
|
signal-induced proliferation associated gene 1 |
| chr10_+_80441855 | 0.05 |
ENSMUST00000178231.2
|
Gm49322
|
predicted gene, 49322 |
| chr11_+_11414256 | 0.05 |
ENSMUST00000020410.11
|
Spata48
|
spermatogenesis associated 48 |
| chr8_-_4267260 | 0.05 |
ENSMUST00000168386.9
|
Prr36
|
proline rich 36 |
| chr8_+_47439916 | 0.05 |
ENSMUST00000039840.15
|
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr9_-_39920878 | 0.05 |
ENSMUST00000216463.3
|
Olfr980
|
olfactory receptor 980 |
| chr16_-_55755208 | 0.05 |
ENSMUST00000121129.8
ENSMUST00000023270.14 |
Cep97
|
centrosomal protein 97 |
| chr5_-_134589025 | 0.05 |
ENSMUST00000149604.5
|
Syna
|
syncytin a |
| chr3_+_88742538 | 0.05 |
ENSMUST00000107498.9
|
Gon4l
|
gon-4-like (C.elegans) |
| chr10_-_95678748 | 0.04 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr10_-_95678786 | 0.04 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chrX_+_100473161 | 0.04 |
ENSMUST00000033673.7
|
Nono
|
non-POU-domain-containing, octamer binding protein |
| chr1_+_65225886 | 0.03 |
ENSMUST00000097707.5
|
Pikfyve
|
phosphoinositide kinase, FYVE type zinc finger containing |
| chr8_-_110688716 | 0.03 |
ENSMUST00000001722.14
ENSMUST00000051430.7 |
Marveld3
|
MARVEL (membrane-associating) domain containing 3 |
| chr11_-_66943389 | 0.02 |
ENSMUST00000116363.2
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese dependent |
| chr6_+_70549568 | 0.02 |
ENSMUST00000196940.2
ENSMUST00000103397.3 |
Igkv3-10
|
immunoglobulin kappa variable 3-10 |
| chr16_+_22877000 | 0.02 |
ENSMUST00000039492.14
ENSMUST00000023589.15 ENSMUST00000089902.8 |
Kng1
|
kininogen 1 |
| chr13_-_59879548 | 0.02 |
ENSMUST00000052978.6
|
Spata31d1d
|
spermatogenesis associated 31 subfamily D, member 1D |
| chr6_+_42222841 | 0.02 |
ENSMUST00000031897.8
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr11_+_120499295 | 0.02 |
ENSMUST00000106194.8
ENSMUST00000106195.3 ENSMUST00000061309.5 |
Npb
|
neuropeptide B |
| chr17_-_74257164 | 0.02 |
ENSMUST00000024866.6
|
Xdh
|
xanthine dehydrogenase |
| chr4_+_154954042 | 0.02 |
ENSMUST00000079269.14
ENSMUST00000163732.8 ENSMUST00000080559.13 |
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr3_+_82933383 | 0.02 |
ENSMUST00000029630.15
ENSMUST00000166581.4 |
Fga
|
fibrinogen alpha chain |
| chr10_-_106959444 | 0.02 |
ENSMUST00000165067.9
|
Acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr2_+_82773971 | 0.01 |
ENSMUST00000143764.9
|
Fsip2
|
fibrous sheath-interacting protein 2 |
| chr16_-_22847829 | 0.01 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr2_+_55325931 | 0.01 |
ENSMUST00000067101.10
|
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr7_+_6463510 | 0.01 |
ENSMUST00000056120.5
|
Olfr1336
|
olfactory receptor 1336 |
| chrY_-_79161056 | 0.01 |
ENSMUST00000179922.2
|
Gm20917
|
predicted gene, 20917 |
| chr5_-_50216249 | 0.01 |
ENSMUST00000030971.7
|
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chr14_+_55170152 | 0.01 |
ENSMUST00000037863.6
|
Il25
|
interleukin 25 |
| chr1_+_21419819 | 0.01 |
ENSMUST00000088407.4
|
Khdc1a
|
KH domain containing 1A |
| chr10_+_33733706 | 0.01 |
ENSMUST00000218204.2
|
Sult3a1
|
sulfotransferase family 3A, member 1 |
| chr10_+_123099945 | 0.01 |
ENSMUST00000238972.2
ENSMUST00000050756.8 |
Tafa2
|
TAFA chemokine like family member 2 |
| chr6_+_126916919 | 0.01 |
ENSMUST00000032497.7
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chrX_-_6907858 | 0.01 |
ENSMUST00000115752.8
|
Ccnb3
|
cyclin B3 |
| chr7_+_28563255 | 0.01 |
ENSMUST00000138272.8
|
Lgals7
|
lectin, galactose binding, soluble 7 |
| chr9_+_104424466 | 0.01 |
ENSMUST00000098443.9
|
Cpne4
|
copine IV |
| chrX_-_101909959 | 0.01 |
ENSMUST00000048061.13
|
1700031F05Rik
|
RIKEN cDNA 1700031F05 gene |
| chr15_+_10358611 | 0.01 |
ENSMUST00000110541.8
ENSMUST00000110542.8 |
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr14_-_50521663 | 0.01 |
ENSMUST00000213701.2
|
Olfr732
|
olfactory receptor 732 |
| chr1_+_21438462 | 0.01 |
ENSMUST00000070223.6
|
Khdc1c
|
KH domain containing 1C |
| chr7_-_6491875 | 0.01 |
ENSMUST00000036357.7
ENSMUST00000220413.2 |
Olfr1347
|
olfactory receptor 1347 |
| chr14_+_53531798 | 0.01 |
ENSMUST00000103626.3
|
Trav9n-4
|
T cell receptor alpha variable 9N-4 |
| chr7_-_19939532 | 0.00 |
ENSMUST00000169774.2
|
Vmn1r241
|
vomeronasal 1 receptor 241 |
| chr7_-_103814019 | 0.00 |
ENSMUST00000154555.2
ENSMUST00000051137.15 |
Olfm5
|
olfactomedin 5 |
| chr17_-_53770324 | 0.00 |
ENSMUST00000024725.5
|
Efhb
|
EF hand domain family, member B |
| chrX_-_91059789 | 0.00 |
ENSMUST00000099471.3
ENSMUST00000072269.2 |
Mageb1
|
MAGE family member B1 |
| chr13_-_105429514 | 0.00 |
ENSMUST00000224011.2
|
Rnf180
|
ring finger protein 180 |
| chrX_+_73171069 | 0.00 |
ENSMUST00000033771.11
ENSMUST00000101457.10 |
Opn1mw
|
opsin 1 (cone pigments), medium-wave-sensitive (color blindness, deutan) |
| chr7_+_23592954 | 0.00 |
ENSMUST00000078593.3
|
Vmn1r178
|
vomeronasal 1 receptor 178 |
| chr11_-_42211868 | 0.00 |
ENSMUST00000020703.13
ENSMUST00000155218.9 |
Gabra6
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 6 |
| chr9_+_75922137 | 0.00 |
ENSMUST00000008052.13
ENSMUST00000183425.8 ENSMUST00000183979.8 ENSMUST00000117981.3 |
Hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase-like 1 |
| chr1_+_5658716 | 0.00 |
ENSMUST00000160777.8
ENSMUST00000239100.2 ENSMUST00000027038.11 |
Oprk1
|
opioid receptor, kappa 1 |
| chr10_-_130116088 | 0.00 |
ENSMUST00000061571.5
|
Neurod4
|
neurogenic differentiation 4 |
| chrY_-_7167777 | 0.00 |
ENSMUST00000178447.2
|
Gm21244
|
predicted gene, 21244 |
| chr16_+_34471558 | 0.00 |
ENSMUST00000023530.5
|
Ropn1
|
ropporin, rhophilin associated protein 1 |
| chr16_+_36695479 | 0.00 |
ENSMUST00000023534.7
ENSMUST00000114812.9 ENSMUST00000134616.8 |
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr17_+_87943401 | 0.00 |
ENSMUST00000235125.2
ENSMUST00000053577.9 ENSMUST00000234009.2 |
Epcam
|
epithelial cell adhesion molecule |
| chr7_-_84059170 | 0.00 |
ENSMUST00000208995.2
|
Arnt2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr7_+_30681287 | 0.00 |
ENSMUST00000128384.3
|
Fam187b
|
family with sequence similarity 187, member B |
| chr2_+_111209312 | 0.00 |
ENSMUST00000209096.2
|
Olfr1284
|
olfactory receptor 1284 |
| chr7_-_115445352 | 0.00 |
ENSMUST00000206369.2
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chr5_+_130398261 | 0.00 |
ENSMUST00000086029.10
|
Caln1
|
calneuron 1 |
| chr16_-_20972750 | 0.00 |
ENSMUST00000170665.3
|
Teddm3
|
transmembrane epididymal family member 3 |
| chr14_-_70761507 | 0.00 |
ENSMUST00000022692.5
|
Sftpc
|
surfactant associated protein C |
| chr11_-_42211886 | 0.00 |
ENSMUST00000109286.4
|
Gabra6
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 6 |
| chr13_-_27574822 | 0.00 |
ENSMUST00000102954.10
ENSMUST00000091678.3 |
Prl2b1
|
prolactin family 2, subfamily b, member 1 |
| chr7_-_19873013 | 0.00 |
ENSMUST00000098747.2
|
Vmn1r93
|
vomeronasal 1 receptor 93 |
| chr15_+_10358643 | 0.00 |
ENSMUST00000022858.8
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr15_-_89263790 | 0.00 |
ENSMUST00000238996.2
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr3_+_107008343 | 0.00 |
ENSMUST00000197470.5
|
Kcna2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr13_-_27622594 | 0.00 |
ENSMUST00000018392.9
ENSMUST00000110355.10 |
Prl8a6
|
prolactin family 8, subfamily a, member 6 |
| chr7_-_115445315 | 0.00 |
ENSMUST00000166207.3
|
Sox6
|
SRY (sex determining region Y)-box 6 |
| chrX_+_111510223 | 0.00 |
ENSMUST00000113409.8
|
Zfp711
|
zinc finger protein 711 |
| chr11_+_49039086 | 0.00 |
ENSMUST00000059379.2
|
Olfr1395
|
olfactory receptor 1395 |
| chr4_+_115268821 | 0.00 |
ENSMUST00000094887.4
|
Cyp4a12b
|
cytochrome P450, family 4, subfamily a, polypeptide 12B |
| chrX_+_77169068 | 0.00 |
ENSMUST00000114030.3
|
Gm5938
|
predicted gene 5938 |
| chr9_+_18946421 | 0.00 |
ENSMUST00000059315.4
|
Olfr835
|
olfactory receptor 835 |
| chr8_-_4266912 | 0.00 |
ENSMUST00000177491.8
|
Prr36
|
proline rich 36 |
| chr17_-_15129766 | 0.00 |
ENSMUST00000228233.2
|
Wdr27
|
WD repeat domain 27 |
| chr6_+_41039255 | 0.00 |
ENSMUST00000103266.3
|
Trbv5
|
T cell receptor beta, variable 5 |
| chr8_-_4267131 | 0.00 |
ENSMUST00000175906.2
|
Prr36
|
proline rich 36 |
| chr13_+_65180860 | 0.00 |
ENSMUST00000099427.3
|
Spata31d1c
|
spermatogenesis associated 31 subfamily D, member 1C |
| chr15_-_98211429 | 0.00 |
ENSMUST00000079736.3
|
Olfr285
|
olfactory receptor 285 |
| chr1_-_90771638 | 0.00 |
ENSMUST00000130846.9
|
Col6a3
|
collagen, type VI, alpha 3 |
| chr11_-_89732091 | 0.00 |
ENSMUST00000238273.3
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr19_-_58781709 | 0.00 |
ENSMUST00000166692.2
|
1700019N19Rik
|
RIKEN cDNA 1700019N19 gene |
| chr2_-_89170692 | 0.00 |
ENSMUST00000099784.5
|
Olfr1233
|
olfactory receptor 1233 |
| chr5_+_138062515 | 0.00 |
ENSMUST00000178402.2
|
Smok3c
|
sperm motility kinase 3C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Aire
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.5 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.2 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.0 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.3 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 1.0 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.6 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.5 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.2 | GO:0070469 | respiratory chain(GO:0070469) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.2 | GO:0050656 | aryl sulfotransferase activity(GO:0004062) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.0 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |