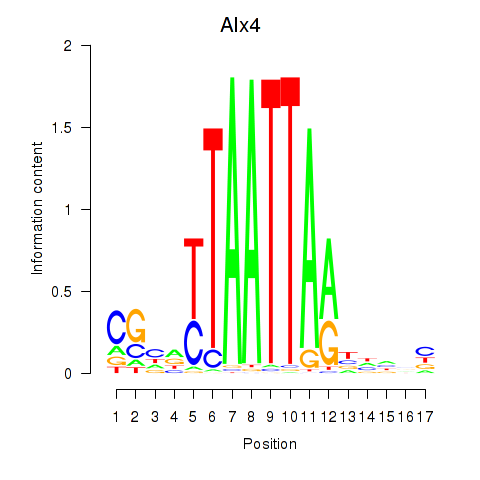
|
chr12_-_75678092
Show fit
|
1.49 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1
|
|
chr5_-_138169253
Show fit
|
1.41 |
ENSMUST00000139983.8
|
Mcm7
|
minichromosome maintenance complex component 7
|
|
chr11_+_87684299
Show fit
|
1.20 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase
|
|
chr3_+_142326363
Show fit
|
1.09 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2
|
|
chr11_+_58062467
Show fit
|
0.95 |
ENSMUST00000020820.2
|
Mrpl22
|
mitochondrial ribosomal protein L22
|
|
chr5_-_138169509
Show fit
|
0.87 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7
|
|
chr13_-_100922910
Show fit
|
0.76 |
ENSMUST00000174038.2
ENSMUST00000091295.14
ENSMUST00000072119.15
|
Ccnb1
|
cyclin B1
|
|
chr8_+_85415935
Show fit
|
0.74 |
ENSMUST00000125370.10
ENSMUST00000175784.2
|
Trmt1
|
tRNA methyltransferase 1
|
|
chr15_-_79658608
Show fit
|
0.73 |
ENSMUST00000229644.2
ENSMUST00000023055.8
|
Dnal4
|
dynein, axonemal, light chain 4
|
|
chr14_-_45626237
Show fit
|
0.70 |
ENSMUST00000227865.2
ENSMUST00000226856.2
ENSMUST00000226276.2
ENSMUST00000046191.9
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr8_-_4829519
Show fit
|
0.68 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1
|
|
chr8_+_31640332
Show fit
|
0.67 |
ENSMUST00000209851.2
ENSMUST00000098842.3
ENSMUST00000210129.2
|
Tti2
|
TELO2 interacting protein 2
|
|
chr7_+_37882642
Show fit
|
0.63 |
ENSMUST00000178207.10
ENSMUST00000179525.10
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene
|
|
chr19_-_24178000
Show fit
|
0.60 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2
|
|
chr8_+_85415876
Show fit
|
0.59 |
ENSMUST00000109767.9
ENSMUST00000177084.8
ENSMUST00000109768.9
ENSMUST00000152301.9
ENSMUST00000177423.8
|
Trmt1
|
tRNA methyltransferase 1
|
|
chr9_-_14292453
Show fit
|
0.59 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1
|
|
chr8_-_85389470
Show fit
|
0.58 |
ENSMUST00000060427.6
|
Ier2
|
immediate early response 2
|
|
chr3_+_89366425
Show fit
|
0.57 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase
|
|
chr1_-_160898181
Show fit
|
0.57 |
ENSMUST00000035430.4
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial)
|
|
chrM_+_8603
Show fit
|
0.57 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III
|
|
chr2_-_29677634
Show fit
|
0.56 |
ENSMUST00000177467.8
ENSMUST00000113807.10
|
Trub2
|
TruB pseudouridine (psi) synthase family member 2
|
|
chr12_-_80690573
Show fit
|
0.55 |
ENSMUST00000166931.2
ENSMUST00000218364.2
|
Erh
|
ERH mRNA splicing and mitosis factor
|
|
chr5_-_65855511
Show fit
|
0.54 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A
|
|
chr19_+_41921903
Show fit
|
0.54 |
ENSMUST00000224258.2
ENSMUST00000026154.9
ENSMUST00000224896.2
|
Zdhhc16
|
zinc finger, DHHC domain containing 16
|
|
chr2_-_144112700
Show fit
|
0.53 |
ENSMUST00000110030.10
|
Snx5
|
sorting nexin 5
|
|
chr11_-_58059293
Show fit
|
0.53 |
ENSMUST00000172035.8
ENSMUST00000035604.13
ENSMUST00000102711.9
|
Gemin5
|
gem nuclear organelle associated protein 5
|
|
chr9_+_21634779
Show fit
|
0.52 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor
|
|
chr11_+_69729340
Show fit
|
0.52 |
ENSMUST00000133967.8
ENSMUST00000094065.5
|
Tmem256
|
transmembrane protein 256
|
|
chr11_+_94632608
Show fit
|
0.51 |
ENSMUST00000021240.7
ENSMUST00000188741.2
|
Cdc34b
|
cell division cycle 34B
|
|
chr12_+_117807224
Show fit
|
0.50 |
ENSMUST00000021592.16
|
Cdca7l
|
cell division cycle associated 7 like
|
|
chr9_+_73009680
Show fit
|
0.49 |
ENSMUST00000034737.13
ENSMUST00000173734.9
ENSMUST00000167514.2
ENSMUST00000174203.3
|
Khdc3
Gm20509
|
KH domain containing 3, subcortical maternal complex member
predicted gene 20509
|
|
chr2_-_131021905
Show fit
|
0.49 |
ENSMUST00000089510.5
|
Cenpb
|
centromere protein B
|
|
chr2_+_174169351
Show fit
|
0.49 |
ENSMUST00000124935.8
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus
|
|
chr6_-_129449739
Show fit
|
0.48 |
ENSMUST00000112076.9
ENSMUST00000184581.3
|
Clec7a
|
C-type lectin domain family 7, member a
|
|
chr4_-_154721288
Show fit
|
0.48 |
ENSMUST00000030902.13
ENSMUST00000105637.8
ENSMUST00000070313.14
ENSMUST00000105636.8
ENSMUST00000105638.9
ENSMUST00000097759.9
ENSMUST00000124771.2
|
Prdm16
|
PR domain containing 16
|
|
chr10_+_128173603
Show fit
|
0.48 |
ENSMUST00000005826.9
|
Cs
|
citrate synthase
|
|
chr15_-_79658584
Show fit
|
0.48 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4
|
|
chr18_-_64649497
Show fit
|
0.46 |
ENSMUST00000237351.2
ENSMUST00000236186.2
ENSMUST00000235325.2
|
Nars
|
asparaginyl-tRNA synthetase
|
|
chr4_-_136329953
Show fit
|
0.44 |
ENSMUST00000105847.8
ENSMUST00000116273.9
|
Kdm1a
|
lysine (K)-specific demethylase 1A
|
|
chr14_+_62529924
Show fit
|
0.43 |
ENSMUST00000166879.8
|
Rnaseh2b
|
ribonuclease H2, subunit B
|
|
chr7_-_110673269
Show fit
|
0.43 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2
|
|
chr7_+_37883074
Show fit
|
0.43 |
ENSMUST00000178876.10
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene
|
|
chr15_+_100202021
Show fit
|
0.43 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1
|
|
chr12_+_10440755
Show fit
|
0.43 |
ENSMUST00000020947.7
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis)
|
|
chr10_-_30076543
Show fit
|
0.42 |
ENSMUST00000099985.6
|
Cenpw
|
centromere protein W
|
|
chr10_-_37014859
Show fit
|
0.42 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate
|
|
chr12_-_79237722
Show fit
|
0.41 |
ENSMUST00000085254.7
|
Rdh11
|
retinol dehydrogenase 11
|
|
chr10_-_128640232
Show fit
|
0.41 |
ENSMUST00000051011.14
|
Tmem198b
|
transmembrane protein 198b
|
|
chr15_+_100202079
Show fit
|
0.40 |
ENSMUST00000230252.2
ENSMUST00000231166.2
|
Mettl7a1
|
methyltransferase like 7A1
|
|
chr5_-_151574620
Show fit
|
0.40 |
ENSMUST00000038131.10
|
Rfc3
|
replication factor C (activator 1) 3
|
|
chr18_-_10030017
Show fit
|
0.39 |
ENSMUST00000116669.2
ENSMUST00000092096.14
|
Usp14
|
ubiquitin specific peptidase 14
|
|
chr4_+_43493344
Show fit
|
0.39 |
ENSMUST00000030181.12
ENSMUST00000107922.3
|
Ccdc107
|
coiled-coil domain containing 107
|
|
chr4_-_135714465
Show fit
|
0.39 |
ENSMUST00000105851.9
|
Pithd1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1
|
|
chr19_-_44095840
Show fit
|
0.38 |
ENSMUST00000119591.2
ENSMUST00000026217.11
|
Chuk
|
conserved helix-loop-helix ubiquitous kinase
|
|
chr9_+_98873831
Show fit
|
0.38 |
ENSMUST00000185472.2
|
Faim
|
Fas apoptotic inhibitory molecule
|
|
chr3_+_89366632
Show fit
|
0.38 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase
|
|
chr1_+_160898283
Show fit
|
0.37 |
ENSMUST00000028035.14
ENSMUST00000111620.10
ENSMUST00000111618.8
|
Cenpl
|
centromere protein L
|
|
chr9_-_22028370
Show fit
|
0.37 |
ENSMUST00000213233.2
|
Elof1
|
ELF1 homolog, elongation factor 1
|
|
chr1_+_190660689
Show fit
|
0.37 |
ENSMUST00000066632.14
ENSMUST00000110899.7
|
Angel2
|
angel homolog 2
|
|
chr11_-_4045343
Show fit
|
0.36 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1
|
|
chrM_+_9459
Show fit
|
0.36 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3
|
|
chrX_-_105264751
Show fit
|
0.35 |
ENSMUST00000113495.9
|
Taf9b
|
TATA-box binding protein associated factor 9B
|
|
chr9_+_65368207
Show fit
|
0.35 |
ENSMUST00000034955.8
ENSMUST00000213957.2
|
Spg21
|
SPG21, maspardin
|
|
chr6_+_11926757
Show fit
|
0.35 |
ENSMUST00000133776.2
|
Phf14
|
PHD finger protein 14
|
|
chr1_+_87983099
Show fit
|
0.34 |
ENSMUST00000138182.8
ENSMUST00000113142.10
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10
|
|
chr8_+_107757847
Show fit
|
0.34 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A
|
|
chr11_-_87249837
Show fit
|
0.33 |
ENSMUST00000055438.5
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing)
|
|
chr7_-_24423715
Show fit
|
0.33 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11
|
|
chr10_+_80003612
Show fit
|
0.33 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein
|
|
chr12_-_102724931
Show fit
|
0.33 |
ENSMUST00000174651.2
|
Gm20604
|
predicted gene 20604
|
|
chr10_-_9776823
Show fit
|
0.33 |
ENSMUST00000141722.8
|
Stxbp5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr7_-_121700958
Show fit
|
0.33 |
ENSMUST00000139456.2
ENSMUST00000106471.9
ENSMUST00000123296.8
ENSMUST00000033157.10
|
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1
|
|
chr14_-_104760051
Show fit
|
0.33 |
ENSMUST00000022716.4
ENSMUST00000228448.2
ENSMUST00000227640.2
|
Obi1
|
ORC ubiquitin ligase 1
|
|
chr6_+_137731599
Show fit
|
0.33 |
ENSMUST00000204356.2
|
Dera
|
deoxyribose-phosphate aldolase (putative)
|
|
chr15_+_100659622
Show fit
|
0.32 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8
|
|
chr1_-_88205233
Show fit
|
0.32 |
ENSMUST00000065420.12
ENSMUST00000054674.15
|
Hjurp
|
Holliday junction recognition protein
|
|
chr7_+_45219766
Show fit
|
0.32 |
ENSMUST00000120864.10
|
Bcat2
|
branched chain aminotransferase 2, mitochondrial
|
|
chr6_+_137731526
Show fit
|
0.32 |
ENSMUST00000203216.3
ENSMUST00000087675.9
ENSMUST00000203693.3
|
Dera
|
deoxyribose-phosphate aldolase (putative)
|
|
chr14_+_55797443
Show fit
|
0.32 |
ENSMUST00000117236.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11
|
|
chr6_-_148847854
Show fit
|
0.31 |
ENSMUST00000139355.8
ENSMUST00000146457.2
ENSMUST00000054080.15
|
Sinhcaf
|
SIN3-HDAC complex associated factor
|
|
chrX_-_94488394
Show fit
|
0.31 |
ENSMUST00000084535.6
|
Amer1
|
APC membrane recruitment 1
|
|
chr1_-_185061525
Show fit
|
0.31 |
ENSMUST00000027921.11
ENSMUST00000110975.8
ENSMUST00000110974.4
|
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial
|
|
chr11_+_106107752
Show fit
|
0.31 |
ENSMUST00000021046.6
|
Ddx42
|
DEAD box helicase 42
|
|
chr3_+_96552895
Show fit
|
0.31 |
ENSMUST00000119365.8
ENSMUST00000029744.6
|
Itga10
|
integrin, alpha 10
|
|
chrX_-_133012600
Show fit
|
0.30 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1
|
|
chr18_+_37898633
Show fit
|
0.30 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12
|
|
chr11_-_79853200
Show fit
|
0.30 |
ENSMUST00000108241.8
ENSMUST00000043152.6
|
Utp6
|
UTP6 small subunit processome component
|
|
chr2_-_126342551
Show fit
|
0.30 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4
|
|
chr18_-_57108405
Show fit
|
0.30 |
ENSMUST00000139243.9
ENSMUST00000025488.15
|
C330018D20Rik
|
RIKEN cDNA C330018D20 gene
|
|
chr16_-_10360893
Show fit
|
0.30 |
ENSMUST00000184863.8
ENSMUST00000038281.6
|
Dexi
|
dexamethasone-induced transcript
|
|
chr13_-_43634695
Show fit
|
0.29 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9
|
|
chr11_-_107228382
Show fit
|
0.29 |
ENSMUST00000040380.13
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr1_+_87983189
Show fit
|
0.29 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10
|
|
chr1_+_87998487
Show fit
|
0.29 |
ENSMUST00000073772.5
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9
|
|
chr4_-_106536063
Show fit
|
0.29 |
ENSMUST00000106772.10
ENSMUST00000135676.2
ENSMUST00000026480.13
|
Ttc4
|
tetratricopeptide repeat domain 4
|
|
chr15_+_65682066
Show fit
|
0.28 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A
|
|
chr1_-_171854818
Show fit
|
0.28 |
ENSMUST00000138714.2
ENSMUST00000027837.13
ENSMUST00000111264.8
|
Vangl2
|
VANGL planar cell polarity 2
|
|
chr1_-_63215812
Show fit
|
0.28 |
ENSMUST00000185847.2
ENSMUST00000185732.7
ENSMUST00000188370.7
ENSMUST00000168099.9
|
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1
|
|
chr5_-_118293443
Show fit
|
0.28 |
ENSMUST00000049474.9
|
Fbxw8
|
F-box and WD-40 domain protein 8
|
|
chr4_+_102446883
Show fit
|
0.28 |
ENSMUST00000097949.11
ENSMUST00000106901.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific
|
|
chr5_+_115417725
Show fit
|
0.27 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase
|
|
chr15_+_102234802
Show fit
|
0.27 |
ENSMUST00000169637.8
|
Pfdn5
|
prefoldin 5
|
|
chr9_-_56151334
Show fit
|
0.27 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1
|
|
chr12_-_79054050
Show fit
|
0.26 |
ENSMUST00000056660.13
ENSMUST00000174721.8
|
Tmem229b
|
transmembrane protein 229B
|
|
chr14_-_59602859
Show fit
|
0.26 |
ENSMUST00000161031.2
|
Phf11d
|
PHD finger protein 11D
|
|
chr11_-_106205320
Show fit
|
0.26 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen
|
|
chr2_+_91376650
Show fit
|
0.26 |
ENSMUST00000099716.11
ENSMUST00000046769.16
ENSMUST00000111337.3
|
Ckap5
|
cytoskeleton associated protein 5
|
|
chr9_-_20871081
Show fit
|
0.26 |
ENSMUST00000177754.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1
|
|
chr17_-_33252341
Show fit
|
0.26 |
ENSMUST00000087654.5
|
Zfp763
|
zinc finger protein 763
|
|
chr14_-_64654397
Show fit
|
0.26 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A
|
|
chr13_+_76727787
Show fit
|
0.25 |
ENSMUST00000126960.8
ENSMUST00000109583.9
|
Mctp1
|
multiple C2 domains, transmembrane 1
|
|
chrX_+_133587268
Show fit
|
0.25 |
ENSMUST00000124226.3
|
Armcx4
|
armadillo repeat containing, X-linked 4
|
|
chr7_+_126832399
Show fit
|
0.25 |
ENSMUST00000056232.7
|
Zfp553
|
zinc finger protein 553
|
|
chr3_+_32762656
Show fit
|
0.24 |
ENSMUST00000029214.14
|
Actl6a
|
actin-like 6A
|
|
chr5_-_23821523
Show fit
|
0.24 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2
|
|
chr4_-_9643636
Show fit
|
0.24 |
ENSMUST00000108333.8
ENSMUST00000108334.8
ENSMUST00000108335.8
ENSMUST00000152526.8
ENSMUST00000103004.10
|
Asph
|
aspartate-beta-hydroxylase
|
|
chr12_+_80690985
Show fit
|
0.24 |
ENSMUST00000219405.2
ENSMUST00000085245.7
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9
|
|
chr12_+_55350023
Show fit
|
0.24 |
ENSMUST00000184766.8
ENSMUST00000183475.8
ENSMUST00000183654.2
|
Prorp
|
protein only RNase P catalytic subunit
|
|
chr7_+_101545547
Show fit
|
0.24 |
ENSMUST00000035395.14
ENSMUST00000106973.8
ENSMUST00000144207.9
|
Anapc15
|
anaphase promoting complex C subunit 15
|
|
chr8_-_35432783
Show fit
|
0.24 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase
|
|
chr11_-_51497665
Show fit
|
0.23 |
ENSMUST00000074669.10
ENSMUST00000101249.9
ENSMUST00000109103.4
|
Hnrnpab
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr12_+_87204374
Show fit
|
0.23 |
ENSMUST00000222222.2
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase)
|
|
chr6_-_115014777
Show fit
|
0.23 |
ENSMUST00000174848.8
ENSMUST00000032461.12
|
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog
|
|
chr17_+_66418598
Show fit
|
0.23 |
ENSMUST00000116556.4
ENSMUST00000233354.2
|
Washc1
|
WASH complex subunit 1
|
|
chr11_+_109434519
Show fit
|
0.23 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G
|
|
chr1_-_78465479
Show fit
|
0.23 |
ENSMUST00000190441.2
ENSMUST00000170217.8
ENSMUST00000188247.7
ENSMUST00000068333.14
|
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit
|
|
chr6_-_148847633
Show fit
|
0.23 |
ENSMUST00000132696.8
|
Sinhcaf
|
SIN3-HDAC complex associated factor
|
|
chr18_-_36587573
Show fit
|
0.23 |
ENSMUST00000025204.7
ENSMUST00000237792.2
|
Pfdn1
|
prefoldin 1
|
|
chr18_-_64649620
Show fit
|
0.22 |
ENSMUST00000025483.11
ENSMUST00000237400.2
|
Nars
|
asparaginyl-tRNA synthetase
|
|
chr2_-_87504008
Show fit
|
0.22 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135
|
|
chr12_+_88327305
Show fit
|
0.22 |
ENSMUST00000101165.9
|
Adck1
|
aarF domain containing kinase 1
|
|
chr7_-_10488291
Show fit
|
0.22 |
ENSMUST00000226874.2
ENSMUST00000227003.2
ENSMUST00000228561.2
ENSMUST00000228248.2
ENSMUST00000228526.2
ENSMUST00000228098.2
ENSMUST00000227940.2
ENSMUST00000228374.2
ENSMUST00000227702.2
|
Vmn1r71
|
vomeronasal 1 receptor 71
|
|
chr7_+_101546059
Show fit
|
0.22 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15
|
|
chr5_-_120750623
Show fit
|
0.22 |
ENSMUST00000140554.2
ENSMUST00000031599.9
ENSMUST00000177800.8
|
Rita1
|
RBPJ interacting and tubulin associated 1
|
|
chr16_+_48692976
Show fit
|
0.21 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma
|
|
chr14_+_55797468
Show fit
|
0.21 |
ENSMUST00000147981.2
ENSMUST00000133256.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11
|
|
chr8_-_117809188
Show fit
|
0.21 |
ENSMUST00000109093.9
ENSMUST00000098375.6
|
Pkd1l2
|
polycystic kidney disease 1 like 2
|
|
chr19_+_5012362
Show fit
|
0.21 |
ENSMUST00000236917.2
|
Mrpl11
|
mitochondrial ribosomal protein L11
|
|
chr3_-_88317601
Show fit
|
0.21 |
ENSMUST00000193338.6
ENSMUST00000056370.13
|
Pmf1
|
polyamine-modulated factor 1
|
|
chr16_+_18655318
Show fit
|
0.21 |
ENSMUST00000055413.13
ENSMUST00000123146.8
ENSMUST00000191388.2
|
2510002D24Rik
|
RIKEN cDNA 2510002D24 gene
|
|
chr12_-_114012399
Show fit
|
0.21 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2
|
|
chr7_+_126575781
Show fit
|
0.20 |
ENSMUST00000206450.2
ENSMUST00000205830.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase)
|
|
chr4_+_147216495
Show fit
|
0.20 |
ENSMUST00000084149.10
|
Zfp991
|
zinc finger protein 991
|
|
chrX_-_133501002
Show fit
|
0.20 |
ENSMUST00000153024.2
|
Gla
|
galactosidase, alpha
|
|
chr16_-_35184211
Show fit
|
0.20 |
ENSMUST00000043521.5
|
Sec22a
|
SEC22 homolog A, vesicle trafficking protein
|
|
chr17_+_66418525
Show fit
|
0.20 |
ENSMUST00000072383.14
|
Washc1
|
WASH complex subunit 1
|
|
chrX_+_20529137
Show fit
|
0.20 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr6_-_13607963
Show fit
|
0.20 |
ENSMUST00000031554.9
ENSMUST00000149123.3
|
Tmem168
|
transmembrane protein 168
|
|
chr12_-_55349760
Show fit
|
0.20 |
ENSMUST00000021410.10
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr2_-_131001916
Show fit
|
0.20 |
ENSMUST00000103188.10
ENSMUST00000133602.8
ENSMUST00000028800.12
|
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene
|
|
chr7_+_23969822
Show fit
|
0.19 |
ENSMUST00000108438.10
|
Zfp93
|
zinc finger protein 93
|
|
chr14_-_64654592
Show fit
|
0.19 |
ENSMUST00000210363.2
|
Msra
|
methionine sulfoxide reductase A
|
|
chr17_-_56343531
Show fit
|
0.19 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1
|
|
chr12_+_88327391
Show fit
|
0.19 |
ENSMUST00000222695.2
|
Adck1
|
aarF domain containing kinase 1
|
|
chr2_-_104847360
Show fit
|
0.19 |
ENSMUST00000111110.3
ENSMUST00000028592.12
|
Eif3m
|
eukaryotic translation initiation factor 3, subunit M
|
|
chr11_+_80074677
Show fit
|
0.19 |
ENSMUST00000017839.3
|
Rnf135
|
ring finger protein 135
|
|
chr11_+_60428788
Show fit
|
0.19 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase
|
|
chr18_+_46730765
Show fit
|
0.19 |
ENSMUST00000238168.2
ENSMUST00000078079.11
ENSMUST00000168382.2
ENSMUST00000235849.2
ENSMUST00000235973.2
ENSMUST00000235455.2
ENSMUST00000237478.2
|
Eif1a
|
eukaryotic translation initiation factor 1A
|
|
chr14_+_32043944
Show fit
|
0.19 |
ENSMUST00000022480.8
ENSMUST00000228529.2
|
Ogdhl
|
oxoglutarate dehydrogenase-like
|
|
chr1_-_152262425
Show fit
|
0.19 |
ENSMUST00000015124.15
|
Tsen15
|
tRNA splicing endonuclease subunit 15
|
|
chr1_+_75498162
Show fit
|
0.19 |
ENSMUST00000027414.16
ENSMUST00000113553.2
|
Stk11ip
|
serine/threonine kinase 11 interacting protein
|
|
chr6_-_69282389
Show fit
|
0.19 |
ENSMUST00000103350.3
|
Igkv4-68
|
immunoglobulin kappa variable 4-68
|
|
chr9_+_19828161
Show fit
|
0.18 |
ENSMUST00000217347.2
ENSMUST00000057596.10
|
Olfr77
|
olfactory receptor 77
|
|
chr17_+_29493113
Show fit
|
0.18 |
ENSMUST00000234326.2
ENSMUST00000235117.2
|
BC004004
|
cDNA sequence BC004004
|
|
chr18_+_44960813
Show fit
|
0.18 |
ENSMUST00000037763.11
|
Ythdc2
|
YTH domain containing 2
|
|
chr14_-_66361931
Show fit
|
0.18 |
ENSMUST00000070515.2
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic
|
|
chr11_+_62442502
Show fit
|
0.18 |
ENSMUST00000136938.2
|
Ubb
|
ubiquitin B
|
|
chr6_+_123239076
Show fit
|
0.18 |
ENSMUST00000032240.4
|
Clec4d
|
C-type lectin domain family 4, member d
|
|
chr19_-_46033353
Show fit
|
0.17 |
ENSMUST00000026252.14
ENSMUST00000156585.9
ENSMUST00000185355.7
ENSMUST00000152946.8
|
Ldb1
|
LIM domain binding 1
|
|
chr13_+_51562675
Show fit
|
0.17 |
ENSMUST00000087978.5
|
S1pr3
|
sphingosine-1-phosphate receptor 3
|
|
chr18_+_31742565
Show fit
|
0.17 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene
|
|
chr2_-_34716083
Show fit
|
0.17 |
ENSMUST00000113077.8
ENSMUST00000028220.10
|
Fbxw2
|
F-box and WD-40 domain protein 2
|
|
chr19_+_8713156
Show fit
|
0.17 |
ENSMUST00000210512.2
ENSMUST00000049424.11
|
Wdr74
|
WD repeat domain 74
|
|
chr6_-_108162513
Show fit
|
0.17 |
ENSMUST00000167338.8
ENSMUST00000172188.2
ENSMUST00000032191.16
|
Sumf1
|
sulfatase modifying factor 1
|
|
chrX_+_139857640
Show fit
|
0.17 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase
|
|
chr17_+_79919267
Show fit
|
0.17 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2
|
|
chr1_-_152262339
Show fit
|
0.16 |
ENSMUST00000162371.2
|
Tsen15
|
tRNA splicing endonuclease subunit 15
|
|
chr5_-_137834444
Show fit
|
0.16 |
ENSMUST00000197586.2
|
Pilra
|
paired immunoglobin-like type 2 receptor alpha
|
|
chr17_+_29493049
Show fit
|
0.16 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004
|
|
chr9_+_21437440
Show fit
|
0.16 |
ENSMUST00000086361.12
ENSMUST00000173769.3
|
AB124611
|
cDNA sequence AB124611
|
|
chr7_+_44221791
Show fit
|
0.16 |
ENSMUST00000002274.10
|
Napsa
|
napsin A aspartic peptidase
|
|
chr6_-_28397997
Show fit
|
0.16 |
ENSMUST00000035930.11
|
Zfp800
|
zinc finger protein 800
|
|
chr3_-_108469468
Show fit
|
0.16 |
ENSMUST00000106622.3
|
Tmem167b
|
transmembrane protein 167B
|
|
chr2_+_22785534
Show fit
|
0.16 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1
|
|
chrX_+_36390430
Show fit
|
0.16 |
ENSMUST00000016553.5
|
Nkap
|
NFKB activating protein
|
|
chr15_+_80017315
Show fit
|
0.15 |
ENSMUST00000023050.9
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1
|
|
chr10_-_129107354
Show fit
|
0.15 |
ENSMUST00000204573.3
|
Olfr777
|
olfactory receptor 777
|
|
chr18_+_23885390
Show fit
|
0.15 |
ENSMUST00000170802.8
ENSMUST00000155708.8
ENSMUST00000118826.9
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr14_+_19801333
Show fit
|
0.15 |
ENSMUST00000022340.5
|
Nid2
|
nidogen 2
|
|
chr12_-_85871281
Show fit
|
0.15 |
ENSMUST00000021676.12
ENSMUST00000142331.2
|
Erg28
|
ergosterol biosynthesis 28
|
|
chrX_+_139857688
Show fit
|
0.15 |
ENSMUST00000239541.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase
|
|
chr17_+_35188888
Show fit
|
0.14 |
ENSMUST00000173680.2
|
Gm20481
|
predicted gene 20481
|
|
chr13_-_81859056
Show fit
|
0.14 |
ENSMUST00000161920.2
ENSMUST00000048993.12
|
Polr3g
|
polymerase (RNA) III (DNA directed) polypeptide G
|
|
chr1_-_88205185
Show fit
|
0.14 |
ENSMUST00000147393.2
|
Hjurp
|
Holliday junction recognition protein
|
|
chr2_-_86109346
Show fit
|
0.14 |
ENSMUST00000217294.2
ENSMUST00000217245.2
ENSMUST00000216432.2
|
Olfr1051
|
olfactory receptor 1051
|
|
chr1_-_63215952
Show fit
|
0.14 |
ENSMUST00000185412.7
ENSMUST00000027111.15
ENSMUST00000189664.2
|
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1
|
|
chr4_+_150938376
Show fit
|
0.14 |
ENSMUST00000073600.9
|
Errfi1
|
ERBB receptor feedback inhibitor 1
|
|
chr5_-_138169476
Show fit
|
0.14 |
ENSMUST00000147920.2
|
Mcm7
|
minichromosome maintenance complex component 7
|
|
chr2_+_32496990
Show fit
|
0.14 |
ENSMUST00000095045.9
ENSMUST00000095044.10
ENSMUST00000126636.8
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr11_+_114566257
Show fit
|
0.13 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2
|