Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
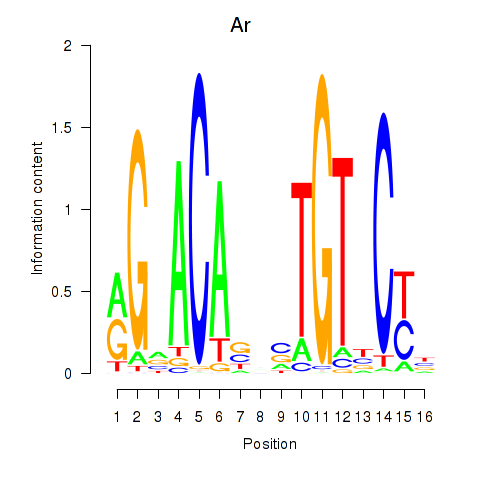
Results for Ar
Z-value: 1.39
Transcription factors associated with Ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ar
|
ENSMUSG00000046532.9 | androgen receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ar | mm39_v1_chrX_+_97192356_97192388 | 0.75 | 1.5e-01 | Click! |
Activity profile of Ar motif
Sorted Z-values of Ar motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_67454476 | 2.33 |
ENSMUST00000049705.8
|
Zfp457
|
zinc finger protein 457 |
| chr9_+_123195986 | 1.46 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr4_-_116565509 | 1.13 |
ENSMUST00000030453.5
|
Mmachc
|
methylmalonic aciduria cblC type, with homocystinuria |
| chr7_-_3551003 | 1.01 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr17_-_23531402 | 0.95 |
ENSMUST00000168033.3
|
Vmn2r114
|
vomeronasal 2, receptor 114 |
| chr16_-_18880821 | 0.94 |
ENSMUST00000200568.2
|
Iglc1
|
immunoglobulin lambda constant 1 |
| chr1_+_172525613 | 0.93 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr8_-_72763462 | 0.85 |
ENSMUST00000003574.5
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr7_+_43701714 | 0.83 |
ENSMUST00000079859.7
|
Klk1b27
|
kallikrein 1-related peptidase b27 |
| chr3_-_122778052 | 0.80 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr7_+_130375799 | 0.78 |
ENSMUST00000048453.7
ENSMUST00000208593.2 |
Btbd16
|
BTB (POZ) domain containing 16 |
| chr11_+_68989763 | 0.75 |
ENSMUST00000021271.14
|
Per1
|
period circadian clock 1 |
| chr13_+_23868175 | 0.74 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr4_-_155729865 | 0.72 |
ENSMUST00000115821.3
|
Gm10563
|
predicted gene 10563 |
| chr18_+_39439778 | 0.70 |
ENSMUST00000235660.2
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr14_-_19600031 | 0.69 |
ENSMUST00000164372.3
|
Gm2244
|
predicted gene 2244 |
| chr15_-_101262452 | 0.68 |
ENSMUST00000230909.2
|
Krt80
|
keratin 80 |
| chrX_-_100838004 | 0.66 |
ENSMUST00000147742.9
|
Gm4779
|
predicted gene 4779 |
| chr7_+_43959637 | 0.65 |
ENSMUST00000107938.8
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr13_-_54836059 | 0.64 |
ENSMUST00000122935.2
ENSMUST00000128257.8 |
Rnf44
|
ring finger protein 44 |
| chr1_+_131936022 | 0.63 |
ENSMUST00000146432.2
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr15_+_64689543 | 0.63 |
ENSMUST00000180105.2
|
Gm21798
|
predicted gene, 21798 |
| chr7_-_85525474 | 0.63 |
ENSMUST00000077478.8
ENSMUST00000233729.2 ENSMUST00000233079.2 ENSMUST00000233315.2 ENSMUST00000233115.2 ENSMUST00000233748.2 ENSMUST00000232965.2 ENSMUST00000232897.2 |
Vmn2r73
|
vomeronasal 2, receptor 73 |
| chr16_+_51852435 | 0.61 |
ENSMUST00000227879.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr7_-_126275529 | 0.58 |
ENSMUST00000106372.11
ENSMUST00000155419.3 ENSMUST00000106373.9 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr17_+_22819932 | 0.58 |
ENSMUST00000097381.5
ENSMUST00000234882.2 |
Vmn2r112
|
vomeronasal 2, receptor 112 |
| chr6_+_122796847 | 0.56 |
ENSMUST00000003238.14
|
Foxj2
|
forkhead box J2 |
| chr11_-_110142565 | 0.55 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr14_+_15018856 | 0.54 |
ENSMUST00000169555.2
|
Gm3755
|
predicted gene 3755 |
| chr17_-_38370970 | 0.53 |
ENSMUST00000216476.2
|
Olfr129
|
olfactory receptor 129 |
| chr14_-_51134930 | 0.51 |
ENSMUST00000227271.2
|
Klhl33
|
kelch-like 33 |
| chr9_+_72892693 | 0.50 |
ENSMUST00000037977.15
|
Ccpg1
|
cell cycle progression 1 |
| chr7_+_16726633 | 0.49 |
ENSMUST00000094805.5
|
Ccdc8
|
coiled-coil domain containing 8 |
| chr17_+_18518361 | 0.48 |
ENSMUST00000231938.2
ENSMUST00000079206.8 ENSMUST00000231879.3 |
Vmn2r93
|
vomeronasal 2, receptor 93 |
| chr17_-_22792463 | 0.48 |
ENSMUST00000092491.7
ENSMUST00000234223.2 ENSMUST00000234296.2 ENSMUST00000234027.2 |
Vmn2r111
|
vomeronasal 2, receptor 111 |
| chr13_-_22225527 | 0.48 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9 |
| chr2_-_59778560 | 0.47 |
ENSMUST00000153136.2
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr5_+_120950690 | 0.46 |
ENSMUST00000086377.9
|
Oas1b
|
2'-5' oligoadenylate synthetase 1B |
| chr11_-_71092282 | 0.44 |
ENSMUST00000108515.9
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr9_+_46910039 | 0.44 |
ENSMUST00000178065.3
|
Gm4791
|
predicted gene 4791 |
| chr2_+_87260619 | 0.44 |
ENSMUST00000215909.2
|
Olfr1124
|
olfactory receptor 1124 |
| chr12_-_103597663 | 0.43 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr5_+_109567554 | 0.43 |
ENSMUST00000232833.2
ENSMUST00000232722.2 ENSMUST00000233536.2 ENSMUST00000171841.3 ENSMUST00000233038.2 ENSMUST00000233724.2 |
Vmn2r17
|
vomeronasal 2, receptor 17 |
| chr15_+_7159038 | 0.43 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr7_-_114162125 | 0.43 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr2_+_83554770 | 0.43 |
ENSMUST00000141725.3
|
Itgav
|
integrin alpha V |
| chr14_+_35816874 | 0.43 |
ENSMUST00000226305.2
|
4930474N05Rik
|
RIKEN cDNA 4930474N05 gene |
| chr12_-_4527138 | 0.43 |
ENSMUST00000085814.5
|
Ncoa1
|
nuclear receptor coactivator 1 |
| chr14_-_17577357 | 0.43 |
ENSMUST00000164459.2
|
Gm3278
|
predicted gene 3278 |
| chr7_-_41708447 | 0.42 |
ENSMUST00000168489.3
ENSMUST00000233456.2 |
Vmn2r59
|
vomeronasal 2, receptor 59 |
| chr13_-_54835996 | 0.42 |
ENSMUST00000150806.8
ENSMUST00000125927.8 |
Rnf44
|
ring finger protein 44 |
| chr11_+_50905063 | 0.41 |
ENSMUST00000217480.2
ENSMUST00000215409.2 |
Olfr54
|
olfactory receptor 54 |
| chr9_+_48406706 | 0.41 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr12_+_84616536 | 0.41 |
ENSMUST00000021665.12
ENSMUST00000169934.4 |
Vsx2
|
visual system homeobox 2 |
| chr11_+_84771133 | 0.40 |
ENSMUST00000020837.7
|
Myo19
|
myosin XIX |
| chr9_+_72892850 | 0.39 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr16_-_92494203 | 0.39 |
ENSMUST00000113956.10
|
Runx1
|
runt related transcription factor 1 |
| chr4_+_150171822 | 0.37 |
ENSMUST00000094451.4
|
Gpr157
|
G protein-coupled receptor 157 |
| chr15_+_54975814 | 0.36 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr5_+_150119860 | 0.36 |
ENSMUST00000202600.4
|
Fry
|
FRY microtubule binding protein |
| chr8_+_73072877 | 0.35 |
ENSMUST00000067912.8
|
Klf2
|
Kruppel-like factor 2 (lung) |
| chr12_+_113112311 | 0.35 |
ENSMUST00000199089.5
|
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr13_-_54835878 | 0.35 |
ENSMUST00000125871.8
|
Rnf44
|
ring finger protein 44 |
| chr7_+_75259778 | 0.35 |
ENSMUST00000207923.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr11_+_105866030 | 0.35 |
ENSMUST00000001964.8
|
Ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr6_+_49013601 | 0.34 |
ENSMUST00000204260.2
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr9_+_110074574 | 0.34 |
ENSMUST00000197850.5
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr3_-_75864195 | 0.33 |
ENSMUST00000038563.14
ENSMUST00000167078.8 ENSMUST00000117242.8 |
Golim4
|
golgi integral membrane protein 4 |
| chr13_-_47196633 | 0.33 |
ENSMUST00000021806.11
ENSMUST00000136864.8 |
Tpmt
|
thiopurine methyltransferase |
| chr11_+_33913013 | 0.33 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr17_+_57071765 | 0.33 |
ENSMUST00000007747.10
|
Dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr7_+_142030744 | 0.33 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr2_-_32277773 | 0.33 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr5_-_135494775 | 0.32 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr5_+_42225303 | 0.32 |
ENSMUST00000087332.5
|
Gm16223
|
predicted gene 16223 |
| chr8_-_116434517 | 0.32 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr11_+_70538083 | 0.32 |
ENSMUST00000037534.8
|
Rnf167
|
ring finger protein 167 |
| chr16_-_95993420 | 0.32 |
ENSMUST00000113804.8
ENSMUST00000054855.14 |
Lca5l
|
Leber congenital amaurosis 5-like |
| chr7_+_97100523 | 0.31 |
ENSMUST00000206658.2
|
Kctd14
|
potassium channel tetramerisation domain containing 14 |
| chr13_+_22656093 | 0.30 |
ENSMUST00000226330.2
ENSMUST00000226965.2 |
Vmn1r201
|
vomeronasal 1 receptor 201 |
| chr9_-_81515865 | 0.30 |
ENSMUST00000183482.2
|
Htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr14_-_19020394 | 0.30 |
ENSMUST00000096168.6
|
Gm10408
|
predicted gene 10408 |
| chrX_-_20955370 | 0.30 |
ENSMUST00000040667.13
|
Zfp300
|
zinc finger protein 300 |
| chr7_-_19415301 | 0.30 |
ENSMUST00000150569.9
ENSMUST00000127648.4 ENSMUST00000003071.10 |
Gm44805
Apoc4
|
predicted gene 44805 apolipoprotein C-IV |
| chr2_+_32617671 | 0.29 |
ENSMUST00000113242.5
|
Sh2d3c
|
SH2 domain containing 3C |
| chr9_+_18315714 | 0.28 |
ENSMUST00000164441.2
ENSMUST00000169398.3 |
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr17_+_18108086 | 0.28 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr14_-_105414681 | 0.27 |
ENSMUST00000163499.2
|
Rbm26
|
RNA binding motif protein 26 |
| chr10_-_7162196 | 0.27 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3 |
| chr3_+_10431961 | 0.27 |
ENSMUST00000029049.7
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr11_-_71092124 | 0.26 |
ENSMUST00000108514.10
|
Nlrp1b
|
NLR family, pyrin domain containing 1B |
| chr17_-_56019571 | 0.26 |
ENSMUST00000086876.7
ENSMUST00000233557.2 ENSMUST00000233972.2 |
Pot1b
|
protection of telomeres 1B |
| chr17_-_35394971 | 0.26 |
ENSMUST00000173324.8
|
Aif1
|
allograft inflammatory factor 1 |
| chr12_-_100865783 | 0.26 |
ENSMUST00000053668.10
|
Gpr68
|
G protein-coupled receptor 68 |
| chr3_-_89820451 | 0.26 |
ENSMUST00000029559.7
ENSMUST00000197679.5 |
Il6ra
|
interleukin 6 receptor, alpha |
| chr12_-_111874489 | 0.26 |
ENSMUST00000054815.15
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr16_+_38279289 | 0.25 |
ENSMUST00000099816.3
ENSMUST00000232409.2 |
Cd80
|
CD80 antigen |
| chr11_-_77678573 | 0.25 |
ENSMUST00000092883.3
|
Gm10277
|
predicted gene 10277 |
| chr5_-_134343491 | 0.24 |
ENSMUST00000173504.8
|
Gtf2i
|
general transcription factor II I |
| chr12_-_111933328 | 0.24 |
ENSMUST00000222874.2
ENSMUST00000223191.2 ENSMUST00000021719.7 |
Atp5mpl
|
ATP synthase membrane subunit 6.8PL |
| chr5_-_23881353 | 0.24 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr7_+_3620356 | 0.24 |
ENSMUST00000076657.11
ENSMUST00000108644.8 |
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr7_+_25318829 | 0.23 |
ENSMUST00000077338.12
ENSMUST00000085953.5 |
Dmac2
|
distal membrane arm assembly complex 2 |
| chr13_-_54836077 | 0.23 |
ENSMUST00000150626.2
ENSMUST00000134177.8 |
Rnf44
|
ring finger protein 44 |
| chr2_-_129139125 | 0.23 |
ENSMUST00000052708.7
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr9_+_98305014 | 0.23 |
ENSMUST00000052068.11
|
Rbp1
|
retinol binding protein 1, cellular |
| chr9_+_72892786 | 0.23 |
ENSMUST00000156879.8
|
Ccpg1
|
cell cycle progression 1 |
| chr10_-_12702674 | 0.22 |
ENSMUST00000219130.2
|
Utrn
|
utrophin |
| chr1_+_89507952 | 0.22 |
ENSMUST00000074945.8
|
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr7_-_4400704 | 0.22 |
ENSMUST00000108590.4
ENSMUST00000206928.2 |
Gp6
|
glycoprotein 6 (platelet) |
| chr4_+_129355374 | 0.22 |
ENSMUST00000048162.10
ENSMUST00000138013.3 |
Bsdc1
|
BSD domain containing 1 |
| chr11_+_96822213 | 0.22 |
ENSMUST00000107633.2
|
Prr15l
|
proline rich 15-like |
| chr18_+_4921663 | 0.22 |
ENSMUST00000143254.8
|
Svil
|
supervillin |
| chr9_+_109865810 | 0.22 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4 |
| chr8_+_75760301 | 0.22 |
ENSMUST00000165630.3
ENSMUST00000212651.2 ENSMUST00000212388.2 ENSMUST00000212299.2 ENSMUST00000078847.13 ENSMUST00000211869.2 |
Tom1
|
target of myb1 trafficking protein |
| chr7_-_85610837 | 0.21 |
ENSMUST00000232886.2
ENSMUST00000232758.2 ENSMUST00000166355.3 |
Vmn2r74
|
vomeronasal 2, receptor 74 |
| chrX_-_36368176 | 0.21 |
ENSMUST00000130324.2
|
Upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr16_-_59138611 | 0.20 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204 |
| chr17_-_24048069 | 0.20 |
ENSMUST00000069579.7
|
Elob
|
elongin B |
| chr17_-_36331416 | 0.20 |
ENSMUST00000174063.2
ENSMUST00000113760.10 |
H2-T24
|
histocompatibility 2, T region locus 24 |
| chr1_-_170417354 | 0.20 |
ENSMUST00000160456.8
|
Nos1ap
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr9_-_123768720 | 0.20 |
ENSMUST00000026911.6
|
Ccr1
|
chemokine (C-C motif) receptor 1 |
| chr1_+_175459735 | 0.19 |
ENSMUST00000097458.4
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr7_+_106630381 | 0.19 |
ENSMUST00000213623.2
|
Olfr713
|
olfactory receptor 713 |
| chr7_-_44741622 | 0.19 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chrX_+_158242121 | 0.19 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr13_-_120496867 | 0.19 |
ENSMUST00000178618.2
|
Gm21188
|
predicted gene, 21188 |
| chrX_-_142716200 | 0.18 |
ENSMUST00000112851.8
ENSMUST00000112856.3 ENSMUST00000033642.10 |
Dcx
|
doublecortin |
| chr15_-_103161237 | 0.18 |
ENSMUST00000154510.8
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr6_+_49013517 | 0.18 |
ENSMUST00000031840.10
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr6_-_129428869 | 0.17 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr10_-_14593935 | 0.17 |
ENSMUST00000020016.5
|
Gje1
|
gap junction protein, epsilon 1 |
| chr9_+_39556381 | 0.17 |
ENSMUST00000219295.2
|
Olfr961
|
olfactory receptor 961 |
| chr9_-_107474221 | 0.17 |
ENSMUST00000238519.2
|
Lsmem2
|
leucine-rich single-pass membrane protein 2 |
| chr2_+_172314433 | 0.17 |
ENSMUST00000029007.3
|
Fam209
|
family with sequence similarity 209 |
| chr7_-_104677667 | 0.17 |
ENSMUST00000215899.2
ENSMUST00000214318.3 |
Olfr675
|
olfactory receptor 675 |
| chr11_-_104333059 | 0.17 |
ENSMUST00000106977.8
ENSMUST00000106972.8 |
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr2_-_91014163 | 0.17 |
ENSMUST00000077941.13
ENSMUST00000111381.9 ENSMUST00000111372.8 ENSMUST00000111371.8 ENSMUST00000075269.10 ENSMUST00000066473.12 |
Madd
|
MAP-kinase activating death domain |
| chr14_-_105414714 | 0.17 |
ENSMUST00000100327.10
ENSMUST00000022715.14 |
Rbm26
|
RNA binding motif protein 26 |
| chr1_-_173707677 | 0.16 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chr8_+_84874881 | 0.16 |
ENSMUST00000093375.5
|
Brme1
|
break repair meiotic recombinase recruitment factor 1 |
| chr15_-_102433050 | 0.16 |
ENSMUST00000023814.9
|
Npff
|
neuropeptide FF-amide peptide precursor |
| chr17_+_80434874 | 0.16 |
ENSMUST00000039205.11
|
Galm
|
galactose mutarotase |
| chr16_-_18884431 | 0.15 |
ENSMUST00000200235.2
|
Iglc3
|
immunoglobulin lambda constant 3 |
| chr19_+_8814782 | 0.15 |
ENSMUST00000171649.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr2_-_73316053 | 0.15 |
ENSMUST00000102680.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr8_+_73373356 | 0.15 |
ENSMUST00000161254.9
|
Nwd1
|
NACHT and WD repeat domain containing 1 |
| chr7_+_23357741 | 0.15 |
ENSMUST00000038694.9
ENSMUST00000173101.3 |
Vmn1r172
|
vomeronasal 1 receptor 172 |
| chr15_-_41733099 | 0.15 |
ENSMUST00000054742.7
|
Abra
|
actin-binding Rho activating protein |
| chr6_+_136495818 | 0.15 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_-_24652775 | 0.14 |
ENSMUST00000123167.2
ENSMUST00000030799.15 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr8_-_70962972 | 0.14 |
ENSMUST00000140679.8
ENSMUST00000129909.8 ENSMUST00000081940.11 |
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr9_+_88721217 | 0.14 |
ENSMUST00000163255.9
ENSMUST00000186363.2 |
Trim43c
|
tripartite motif-containing 43C |
| chr11_+_9143415 | 0.14 |
ENSMUST00000151522.2
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr8_+_111646548 | 0.14 |
ENSMUST00000117534.8
ENSMUST00000034197.5 |
St3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr17_-_37888554 | 0.14 |
ENSMUST00000216181.2
|
Olfr113
|
olfactory receptor 113 |
| chr7_+_43751749 | 0.14 |
ENSMUST00000085455.6
|
Klk1b21
|
kallikrein 1-related peptidase b21 |
| chr10_-_117118226 | 0.14 |
ENSMUST00000092163.9
|
Lyz2
|
lysozyme 2 |
| chr19_+_11352862 | 0.14 |
ENSMUST00000188995.2
|
Ms4a4a
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr14_-_17283980 | 0.14 |
ENSMUST00000171053.2
|
Gm8271
|
predicted gene 8271 |
| chr8_+_120301974 | 0.14 |
ENSMUST00000093100.3
|
Dnaaf1
|
dynein, axonemal assembly factor 1 |
| chr2_-_36899347 | 0.14 |
ENSMUST00000216437.2
|
Olfr358
|
olfactory receptor 358 |
| chr9_+_21867043 | 0.14 |
ENSMUST00000053583.7
|
Swsap1
|
SWIM type zinc finger 7 associated protein 1 |
| chr3_+_97536120 | 0.14 |
ENSMUST00000107050.8
ENSMUST00000029729.15 ENSMUST00000107049.2 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr15_+_98927736 | 0.14 |
ENSMUST00000058914.10
|
Tuba1c
|
tubulin, alpha 1C |
| chr1_+_88015524 | 0.13 |
ENSMUST00000113139.2
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr13_-_67523832 | 0.13 |
ENSMUST00000225787.2
ENSMUST00000172266.8 ENSMUST00000057070.9 |
Zfp456
|
zinc finger protein 456 |
| chr11_-_46581135 | 0.13 |
ENSMUST00000169584.8
|
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr18_+_64387428 | 0.12 |
ENSMUST00000025477.15
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr6_-_125143425 | 0.12 |
ENSMUST00000117757.9
ENSMUST00000073605.15 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr17_-_79328157 | 0.12 |
ENSMUST00000168887.8
ENSMUST00000119284.8 |
Prkd3
|
protein kinase D3 |
| chr14_+_30201569 | 0.12 |
ENSMUST00000022535.9
ENSMUST00000223658.2 |
Dcp1a
|
decapping mRNA 1A |
| chr7_-_82297676 | 0.12 |
ENSMUST00000207693.2
ENSMUST00000056728.5 ENSMUST00000156720.8 ENSMUST00000126478.2 |
Saxo2
|
stabilizer of axonemal microtubules 2 |
| chr17_-_56381540 | 0.12 |
ENSMUST00000139371.2
|
Ubxn6
|
UBX domain protein 6 |
| chr16_+_13176238 | 0.12 |
ENSMUST00000149359.2
|
Mrtfb
|
myocardin related transcription factor B |
| chr11_+_101443674 | 0.12 |
ENSMUST00000107213.8
ENSMUST00000107208.8 ENSMUST00000107212.8 ENSMUST00000127421.8 |
Nbr1
|
NBR1, autophagy cargo receptor |
| chr10_-_12424623 | 0.12 |
ENSMUST00000219003.2
|
Utrn
|
utrophin |
| chr15_-_79287747 | 0.12 |
ENSMUST00000074991.10
|
Tmem184b
|
transmembrane protein 184b |
| chr11_+_69016722 | 0.12 |
ENSMUST00000021268.9
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr7_-_137012444 | 0.12 |
ENSMUST00000120340.2
ENSMUST00000117404.8 ENSMUST00000068996.13 |
9430038I01Rik
|
RIKEN cDNA 9430038I01 gene |
| chr11_+_94827050 | 0.11 |
ENSMUST00000001547.8
|
Col1a1
|
collagen, type I, alpha 1 |
| chr11_+_58549846 | 0.11 |
ENSMUST00000213232.2
|
Olfr322
|
olfactory receptor 322 |
| chr10_-_128016135 | 0.11 |
ENSMUST00000238843.2
ENSMUST00000099139.9 |
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr10_-_33500583 | 0.11 |
ENSMUST00000161692.2
ENSMUST00000160299.2 ENSMUST00000019920.13 |
Clvs2
|
clavesin 2 |
| chr13_+_84370405 | 0.11 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chrX_+_73298388 | 0.11 |
ENSMUST00000119197.8
ENSMUST00000088313.5 |
Emd
|
emerin |
| chr7_-_99345016 | 0.11 |
ENSMUST00000107086.9
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr3_+_90561560 | 0.11 |
ENSMUST00000079286.4
|
S100a7a
|
S100 calcium binding protein A7A |
| chr15_+_100202642 | 0.11 |
ENSMUST00000067752.5
ENSMUST00000229588.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr10_+_79824418 | 0.11 |
ENSMUST00000004784.11
ENSMUST00000105374.2 |
Cnn2
|
calponin 2 |
| chr4_+_108576846 | 0.11 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr11_-_75330302 | 0.11 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chrX_+_20483742 | 0.11 |
ENSMUST00000115375.8
ENSMUST00000115374.8 ENSMUST00000084383.10 |
Rbm10
|
RNA binding motif protein 10 |
| chr7_+_6386294 | 0.11 |
ENSMUST00000081022.9
|
Zfp28
|
zinc finger protein 28 |
| chr18_-_61344452 | 0.11 |
ENSMUST00000148829.2
|
Slc26a2
|
solute carrier family 26 (sulfate transporter), member 2 |
| chr1_-_167070695 | 0.11 |
ENSMUST00000027839.14
|
Uck2
|
uridine-cytidine kinase 2 |
| chr5_+_122534900 | 0.11 |
ENSMUST00000196969.5
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr7_+_82297803 | 0.10 |
ENSMUST00000141726.8
ENSMUST00000179489.8 ENSMUST00000039881.4 |
Efl1
|
elongation factor like GTPase 1 |
| chr2_+_31578537 | 0.10 |
ENSMUST00000075759.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr1_-_133728779 | 0.10 |
ENSMUST00000143567.8
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ar
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.4 | 2.3 | GO:0070893 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.3 | 0.8 | GO:0009233 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 1.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.5 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 0.7 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.1 | 1.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.8 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 0.3 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.1 | 0.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.4 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.1 | 0.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.3 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.3 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.3 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.1 | 0.3 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.7 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 | 0.4 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) regulation of response to drug(GO:2001023) |
| 0.1 | 0.7 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.6 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.3 | GO:0014063 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 0.2 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.1 | 0.3 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.4 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.3 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.5 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:2000536 | negative regulation of entry of bacterium into host cell(GO:2000536) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) UMP salvage(GO:0044206) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:1990051 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.1 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.0 | 0.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.4 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.4 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.0 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0034683 | integrin alphav-beta3 complex(GO:0034683) integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.3 | 0.8 | GO:0052870 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.1 | 0.4 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.1 | 0.6 | GO:0050656 | aryl sulfotransferase activity(GO:0004062) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.7 | GO:0071532 | GKAP/Homer scaffold activity(GO:0030160) ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.3 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.1 | 0.3 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |