Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
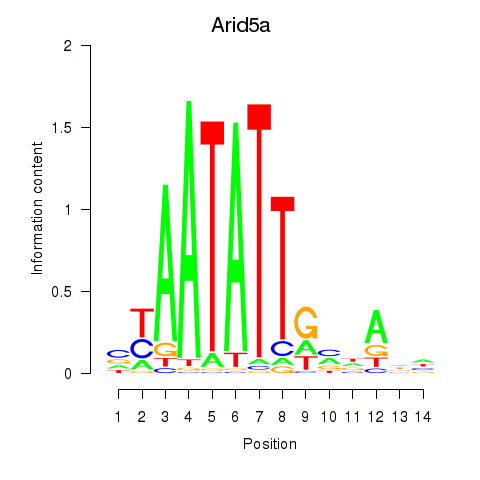
Results for Arid5a
Z-value: 0.54
Transcription factors associated with Arid5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arid5a
|
ENSMUSG00000037447.17 | AT rich interactive domain 5A (MRF1-like) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arid5a | mm39_v1_chr1_+_36346824_36346850 | -0.46 | 4.3e-01 | Click! |
Activity profile of Arid5a motif
Sorted Z-values of Arid5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_115198093 | 0.33 |
ENSMUST00000219890.2
ENSMUST00000218731.2 ENSMUST00000217887.2 ENSMUST00000092170.7 |
Tmem19
|
transmembrane protein 19 |
| chrM_+_8603 | 0.33 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr13_+_94954202 | 0.28 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr10_-_115197775 | 0.25 |
ENSMUST00000217848.2
|
Tmem19
|
transmembrane protein 19 |
| chr17_-_35265702 | 0.23 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chr15_-_55421144 | 0.22 |
ENSMUST00000172387.8
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chrM_+_7758 | 0.22 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr15_+_41573995 | 0.20 |
ENSMUST00000229769.2
|
Oxr1
|
oxidation resistance 1 |
| chr3_-_146487102 | 0.18 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr11_+_98689479 | 0.17 |
ENSMUST00000037930.13
|
Msl1
|
male specific lethal 1 |
| chrM_+_7779 | 0.15 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chrX_-_133012600 | 0.13 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr9_+_96140781 | 0.09 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr17_-_37594351 | 0.05 |
ENSMUST00000216328.2
|
Olfr99
|
olfactory receptor 99 |
| chr9_+_96140750 | 0.04 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr3_+_75982890 | 0.02 |
ENSMUST00000160261.8
|
Fstl5
|
follistatin-like 5 |
| chr5_-_87074380 | 0.02 |
ENSMUST00000031183.3
|
Ugt2b1
|
UDP glucuronosyltransferase 2 family, polypeptide B1 |
| chr10_+_52267702 | 0.02 |
ENSMUST00000067085.7
|
Nepn
|
nephrocan |
| chr4_-_52919172 | 0.01 |
ENSMUST00000107667.3
ENSMUST00000213989.2 |
Olfr272
|
olfactory receptor 272 |
| chr4_+_43384320 | 0.01 |
ENSMUST00000136360.2
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr2_+_61876806 | 0.01 |
ENSMUST00000102735.10
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chrX_-_133012457 | 0.01 |
ENSMUST00000159259.3
ENSMUST00000113275.10 |
Nox1
|
NADPH oxidase 1 |
| chr10_+_69370038 | 0.01 |
ENSMUST00000182439.8
ENSMUST00000092434.12 ENSMUST00000047061.13 ENSMUST00000092432.12 ENSMUST00000092431.12 ENSMUST00000054167.15 |
Ank3
|
ankyrin 3, epithelial |
| chr18_+_37637317 | 0.01 |
ENSMUST00000052179.8
|
Pcdhb20
|
protocadherin beta 20 |
| chr2_+_105505823 | 0.01 |
ENSMUST00000167211.9
ENSMUST00000111083.10 |
Pax6
|
paired box 6 |
| chr1_-_162913210 | 0.01 |
ENSMUST00000096608.5
|
Mroh9
|
maestro heat-like repeat family member 9 |
| chr9_+_37996134 | 0.01 |
ENSMUST00000074681.2
|
Olfr887
|
olfactory receptor 887 |
| chr1_+_133965228 | 0.01 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr2_+_58644922 | 0.01 |
ENSMUST00000059102.13
|
Upp2
|
uridine phosphorylase 2 |
| chr2_+_61876923 | 0.01 |
ENSMUST00000054484.15
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_+_61876956 | 0.01 |
ENSMUST00000112480.3
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 |
| chr2_+_58645189 | 0.01 |
ENSMUST00000102755.4
ENSMUST00000230627.2 ENSMUST00000229923.2 |
Upp2
|
uridine phosphorylase 2 |
| chr10_+_129298547 | 0.01 |
ENSMUST00000077836.3
|
Olfr787
|
olfactory receptor 787 |
| chr2_-_111476340 | 0.00 |
ENSMUST00000119566.4
|
Olfr1298
|
olfactory receptor 1298 |
| chr7_-_105249308 | 0.00 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr2_-_79959802 | 0.00 |
ENSMUST00000102653.8
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr5_+_68189116 | 0.00 |
ENSMUST00000094715.8
|
Grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr5_-_18054702 | 0.00 |
ENSMUST00000165232.8
|
Cd36
|
CD36 molecule |
| chr7_-_132616977 | 0.00 |
ENSMUST00000169570.8
|
Ctbp2
|
C-terminal binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arid5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0072592 | positive regulation of integrin biosynthetic process(GO:0045726) oxygen metabolic process(GO:0072592) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |