Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
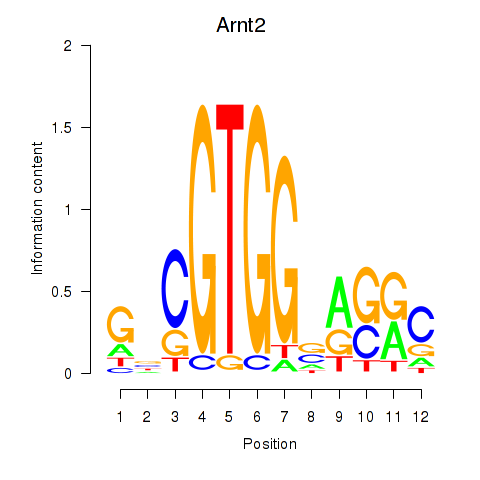
Results for Arnt2
Z-value: 0.34
Transcription factors associated with Arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt2
|
ENSMUSG00000015709.10 | aryl hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arnt2 | mm39_v1_chr7_-_84059170_84059210 | 0.36 | 5.5e-01 | Click! |
Activity profile of Arnt2 motif
Sorted Z-values of Arnt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_23736205 | 0.22 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr17_-_23964807 | 0.16 |
ENSMUST00000046525.10
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr15_+_31224555 | 0.16 |
ENSMUST00000186109.2
|
Dap
|
death-associated protein |
| chr11_+_107438751 | 0.14 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr2_-_33261498 | 0.13 |
ENSMUST00000113165.8
|
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr3_+_96604390 | 0.13 |
ENSMUST00000162778.3
ENSMUST00000064900.16 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr2_-_33261411 | 0.13 |
ENSMUST00000131298.7
ENSMUST00000091039.5 ENSMUST00000042615.13 |
Ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr1_+_59952131 | 0.11 |
ENSMUST00000036540.12
|
Fam117b
|
family with sequence similarity 117, member B |
| chr1_-_52856801 | 0.11 |
ENSMUST00000027271.9
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr6_+_4601124 | 0.11 |
ENSMUST00000141359.2
|
Casd1
|
CAS1 domain containing 1 |
| chr1_-_59012579 | 0.06 |
ENSMUST00000173590.2
ENSMUST00000027186.12 |
Trak2
|
trafficking protein, kinesin binding 2 |
| chr1_+_59012680 | 0.06 |
ENSMUST00000114296.8
ENSMUST00000027185.11 |
Stradb
|
STE20-related kinase adaptor beta |
| chrX_-_52358663 | 0.05 |
ENSMUST00000114841.2
ENSMUST00000071023.12 |
Fam122b
|
family with sequence similarity 122, member B |
| chr2_+_91480460 | 0.05 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr3_+_96604415 | 0.05 |
ENSMUST00000107077.4
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr15_+_31224616 | 0.04 |
ENSMUST00000186547.7
|
Dap
|
death-associated protein |
| chr2_+_79085844 | 0.04 |
ENSMUST00000099972.5
|
Itga4
|
integrin alpha 4 |
| chr19_-_5148506 | 0.03 |
ENSMUST00000025805.8
|
Cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr7_+_18810167 | 0.02 |
ENSMUST00000108479.2
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr15_+_31224460 | 0.02 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr11_+_43419586 | 0.02 |
ENSMUST00000050574.7
|
Ccnjl
|
cyclin J-like |
| chr5_+_88712840 | 0.02 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr9_+_65278979 | 0.01 |
ENSMUST00000239433.2
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr7_+_18810097 | 0.01 |
ENSMUST00000032570.14
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr4_-_123217391 | 0.01 |
ENSMUST00000102640.2
|
Oxct2a
|
3-oxoacid CoA transferase 2A |
| chr4_-_129534752 | 0.01 |
ENSMUST00000132217.8
ENSMUST00000130017.2 ENSMUST00000154105.8 |
Txlna
|
taxilin alpha |
| chr1_-_52856662 | 0.01 |
ENSMUST00000162576.8
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr9_-_44231526 | 0.01 |
ENSMUST00000214602.2
ENSMUST00000065080.10 |
C2cd2l
|
C2 calcium-dependent domain containing 2-like |
| chr4_+_123010035 | 0.00 |
ENSMUST00000102648.6
|
Oxct2b
|
3-oxoacid CoA transferase 2B |
| chr7_-_44741138 | 0.00 |
ENSMUST00000210527.2
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr7_+_130467564 | 0.00 |
ENSMUST00000075181.11
ENSMUST00000151119.9 ENSMUST00000048180.12 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr2_+_119039456 | 0.00 |
ENSMUST00000102519.5
|
Zfyve19
|
zinc finger, FYVE domain containing 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Arnt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |