Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
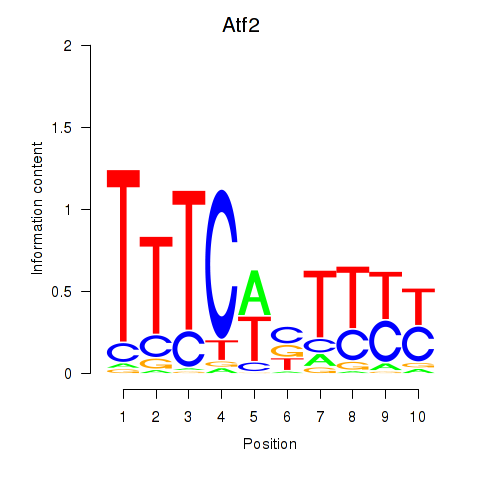
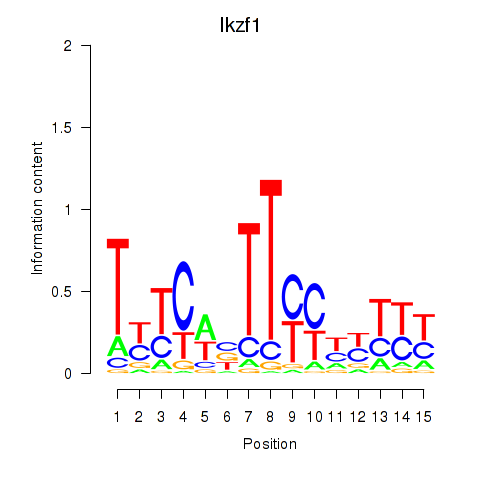
Results for Atf2_Ikzf1
Z-value: 1.55
Transcription factors associated with Atf2_Ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf2
|
ENSMUSG00000027104.19 | activating transcription factor 2 |
|
Ikzf1
|
ENSMUSG00000018654.18 | IKAROS family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf2 | mm39_v1_chr2_-_73722874_73722900 | 0.98 | 2.3e-03 | Click! |
| Ikzf1 | mm39_v1_chr11_+_11635908_11635949 | 0.96 | 7.9e-03 | Click! |
Activity profile of Atf2_Ikzf1 motif
Sorted Z-values of Atf2_Ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23715220 | 1.59 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr13_-_40887244 | 1.44 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr8_+_108669276 | 1.22 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr6_-_25689781 | 1.22 |
ENSMUST00000200812.2
|
Gpr37
|
G protein-coupled receptor 37 |
| chr2_+_93472657 | 1.17 |
ENSMUST00000042078.10
ENSMUST00000111254.2 |
Alx4
|
aristaless-like homeobox 4 |
| chr3_+_96127174 | 1.10 |
ENSMUST00000073115.5
|
H2ac21
|
H2A clustered histone 21 |
| chr5_+_107655487 | 1.06 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr7_-_3918484 | 0.96 |
ENSMUST00000038176.15
ENSMUST00000206077.2 ENSMUST00000090689.5 |
Lilra6
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 |
| chr14_-_67106037 | 0.92 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr14_+_25459690 | 0.90 |
ENSMUST00000007961.15
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr9_-_91247809 | 0.87 |
ENSMUST00000034927.13
|
Zic1
|
zinc finger protein of the cerebellum 1 |
| chrX_-_133062677 | 0.86 |
ENSMUST00000033611.5
|
Xkrx
|
X-linked Kx blood group related, X-linked |
| chr1_-_71692320 | 0.86 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr13_+_21919225 | 0.84 |
ENSMUST00000087714.6
|
H4c11
|
H4 clustered histone 11 |
| chr16_+_43067641 | 0.83 |
ENSMUST00000079441.13
ENSMUST00000114691.8 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr13_+_99321241 | 0.83 |
ENSMUST00000056558.11
|
Zfp366
|
zinc finger protein 366 |
| chr7_-_106709576 | 0.82 |
ENSMUST00000215713.2
|
Olfr715b
|
olfactory receptor 715B |
| chr13_+_54519161 | 0.80 |
ENSMUST00000026985.9
|
Cplx2
|
complexin 2 |
| chr10_+_51356728 | 0.80 |
ENSMUST00000102894.6
ENSMUST00000219661.2 ENSMUST00000219696.2 ENSMUST00000217706.2 |
Lilr4b
Gm49339
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4B predicted gene, 49339 |
| chr10_+_51367052 | 0.78 |
ENSMUST00000217705.2
ENSMUST00000078778.5 ENSMUST00000220182.2 ENSMUST00000220226.2 |
Lilrb4a
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4A |
| chr18_-_80751327 | 0.72 |
ENSMUST00000236310.2
ENSMUST00000167977.8 ENSMUST00000035800.8 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr14_-_122202508 | 0.71 |
ENSMUST00000227267.2
|
Gpr183
|
G protein-coupled receptor 183 |
| chrX_-_72913410 | 0.71 |
ENSMUST00000066576.12
ENSMUST00000114430.8 |
L1cam
|
L1 cell adhesion molecule |
| chr2_-_90309509 | 0.70 |
ENSMUST00000111495.9
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr10_-_43880353 | 0.68 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chrX_-_104919201 | 0.66 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr5_-_132570710 | 0.63 |
ENSMUST00000182974.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr12_-_80159768 | 0.63 |
ENSMUST00000219642.2
ENSMUST00000165114.2 ENSMUST00000218835.2 ENSMUST00000021552.3 |
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr9_-_54858282 | 0.62 |
ENSMUST00000054018.7
|
AY074887
|
cDNA sequence AY074887 |
| chr4_+_132903646 | 0.62 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr11_-_69649004 | 0.61 |
ENSMUST00000071213.4
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr9_+_119978773 | 0.61 |
ENSMUST00000068698.15
ENSMUST00000215512.2 ENSMUST00000111627.3 ENSMUST00000093773.8 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr13_-_12355604 | 0.60 |
ENSMUST00000168193.8
ENSMUST00000064204.14 |
Actn2
|
actinin alpha 2 |
| chr11_+_98818640 | 0.58 |
ENSMUST00000107474.8
|
Rara
|
retinoic acid receptor, alpha |
| chr9_+_27702243 | 0.55 |
ENSMUST00000115243.9
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr14_-_122202599 | 0.55 |
ENSMUST00000049872.9
|
Gpr183
|
G protein-coupled receptor 183 |
| chr4_+_150366028 | 0.54 |
ENSMUST00000105682.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr10_+_127256736 | 0.54 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chr13_+_41071077 | 0.53 |
ENSMUST00000067778.8
ENSMUST00000225759.2 |
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr11_-_18968979 | 0.52 |
ENSMUST00000144988.8
|
Meis1
|
Meis homeobox 1 |
| chr2_+_4305273 | 0.50 |
ENSMUST00000175669.8
|
Frmd4a
|
FERM domain containing 4A |
| chr15_-_50752437 | 0.49 |
ENSMUST00000183997.8
ENSMUST00000183757.8 |
Trps1
|
transcriptional repressor GATA binding 1 |
| chrX_+_84091915 | 0.49 |
ENSMUST00000239019.2
ENSMUST00000113992.3 ENSMUST00000113991.8 |
Dmd
|
dystrophin, muscular dystrophy |
| chr9_-_106353303 | 0.49 |
ENSMUST00000156426.8
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr12_+_116239006 | 0.48 |
ENSMUST00000090195.5
|
Gm11027
|
predicted gene 11027 |
| chr7_-_80051455 | 0.48 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr11_-_18968955 | 0.48 |
ENSMUST00000068264.14
ENSMUST00000185131.8 |
Meis1
|
Meis homeobox 1 |
| chr10_-_10958031 | 0.47 |
ENSMUST00000105561.9
ENSMUST00000044306.13 |
Grm1
|
glutamate receptor, metabotropic 1 |
| chr11_-_109188917 | 0.46 |
ENSMUST00000106704.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr4_-_24800890 | 0.46 |
ENSMUST00000108214.9
|
Klhl32
|
kelch-like 32 |
| chr1_+_173093568 | 0.45 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418 |
| chr12_+_84820024 | 0.45 |
ENSMUST00000021667.7
ENSMUST00000222449.2 ENSMUST00000222982.2 |
Isca2
|
iron-sulfur cluster assembly 2 |
| chr14_-_33700719 | 0.45 |
ENSMUST00000166737.2
|
Zfp488
|
zinc finger protein 488 |
| chr11_-_69649452 | 0.44 |
ENSMUST00000058470.16
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr11_-_97590460 | 0.44 |
ENSMUST00000103148.8
ENSMUST00000169807.8 |
Pcgf2
|
polycomb group ring finger 2 |
| chr13_+_23317325 | 0.44 |
ENSMUST00000227050.2
ENSMUST00000227160.2 ENSMUST00000227741.2 ENSMUST00000226692.2 |
Vmn1r218
|
vomeronasal 1 receptor 218 |
| chr10_-_126877382 | 0.43 |
ENSMUST00000116231.4
|
Eef1akmt3
|
EEF1A lysine methyltransferase 3 |
| chr5_-_142892457 | 0.42 |
ENSMUST00000167721.8
ENSMUST00000163829.2 ENSMUST00000100497.11 |
Actb
|
actin, beta |
| chr19_+_26600820 | 0.42 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_-_48474356 | 0.42 |
ENSMUST00000027764.10
ENSMUST00000053612.14 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr2_-_165242307 | 0.42 |
ENSMUST00000029213.5
|
Ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr3_+_96484294 | 0.41 |
ENSMUST00000148290.2
|
Gm16253
|
predicted gene 16253 |
| chr15_+_82159094 | 0.40 |
ENSMUST00000116423.3
ENSMUST00000230418.2 |
Septin3
|
septin 3 |
| chr10_-_129524028 | 0.40 |
ENSMUST00000203785.3
ENSMUST00000217576.2 |
Olfr802
|
olfactory receptor 802 |
| chr11_+_77873535 | 0.40 |
ENSMUST00000108360.8
ENSMUST00000049167.14 |
Phf12
|
PHD finger protein 12 |
| chr7_+_75351713 | 0.40 |
ENSMUST00000207239.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr10_-_78688000 | 0.39 |
ENSMUST00000205100.3
|
Olfr1356
|
olfactory receptor 1356 |
| chr18_+_36481706 | 0.39 |
ENSMUST00000235864.2
ENSMUST00000050584.10 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr9_+_38053813 | 0.39 |
ENSMUST00000213458.2
|
Olfr890
|
olfactory receptor 890 |
| chr11_+_70453724 | 0.38 |
ENSMUST00000102559.11
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr6_-_38852857 | 0.38 |
ENSMUST00000162359.8
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr11_+_29413734 | 0.38 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr2_-_37320848 | 0.38 |
ENSMUST00000053098.6
|
Zbtb6
|
zinc finger and BTB domain containing 6 |
| chr11_+_70453666 | 0.38 |
ENSMUST00000072237.13
ENSMUST00000072873.14 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr18_+_69726654 | 0.36 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chr15_-_102630589 | 0.36 |
ENSMUST00000023818.11
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr11_-_18968714 | 0.36 |
ENSMUST00000177417.8
|
Meis1
|
Meis homeobox 1 |
| chr7_-_29204812 | 0.36 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr7_-_15781838 | 0.35 |
ENSMUST00000210781.2
|
Bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr11_-_78950698 | 0.35 |
ENSMUST00000141409.8
|
Ksr1
|
kinase suppressor of ras 1 |
| chr4_+_140633646 | 0.35 |
ENSMUST00000030765.7
|
Padi2
|
peptidyl arginine deiminase, type II |
| chr7_+_79676095 | 0.35 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
| chr7_-_106605189 | 0.35 |
ENSMUST00000216375.2
ENSMUST00000208147.3 |
Olfr2
|
olfactory receptor 2 |
| chr8_-_73228953 | 0.35 |
ENSMUST00000079510.6
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr7_+_126184108 | 0.35 |
ENSMUST00000039522.8
|
Apobr
|
apolipoprotein B receptor |
| chr6_+_129489501 | 0.35 |
ENSMUST00000032263.7
|
Tmem52b
|
transmembrane protein 52B |
| chr17_+_48623157 | 0.34 |
ENSMUST00000049614.13
|
B430306N03Rik
|
RIKEN cDNA B430306N03 gene |
| chr16_+_33504740 | 0.34 |
ENSMUST00000232568.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr9_+_44151962 | 0.34 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr8_-_85500010 | 0.34 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr2_-_73284262 | 0.34 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr6_+_136509922 | 0.34 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr14_-_56121824 | 0.33 |
ENSMUST00000063871.13
|
Cbln3
|
cerebellin 3 precursor protein |
| chr13_+_49658249 | 0.33 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chrX_-_72830487 | 0.33 |
ENSMUST00000052761.9
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD+), gamma |
| chr13_+_54931800 | 0.33 |
ENSMUST00000142158.8
ENSMUST00000139184.8 ENSMUST00000132415.8 ENSMUST00000132005.8 ENSMUST00000126785.8 ENSMUST00000141398.2 ENSMUST00000132728.8 ENSMUST00000110003.9 ENSMUST00000152204.3 |
Eif4e1b
|
eukaryotic translation initiation factor 4E family member 1B |
| chr9_+_106306598 | 0.33 |
ENSMUST00000150576.8
|
Rpl29
|
ribosomal protein L29 |
| chr7_+_142014546 | 0.33 |
ENSMUST00000105968.8
ENSMUST00000018963.11 ENSMUST00000105967.8 |
Lsp1
|
lymphocyte specific 1 |
| chr11_-_110058899 | 0.33 |
ENSMUST00000044850.4
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr14_+_51366512 | 0.33 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr3_+_105821450 | 0.33 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr1_-_160134873 | 0.32 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr11_-_78136767 | 0.32 |
ENSMUST00000002121.5
|
Supt6
|
SPT6, histone chaperone and transcription elongation factor |
| chr7_-_102638531 | 0.32 |
ENSMUST00000215606.2
|
Olfr578
|
olfactory receptor 578 |
| chr8_-_116434517 | 0.32 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr9_+_106247943 | 0.31 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chrX_-_9335525 | 0.31 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr7_-_106730290 | 0.31 |
ENSMUST00000214919.2
|
Olfr715
|
olfactory receptor 715 |
| chr8_+_117822593 | 0.31 |
ENSMUST00000034308.16
ENSMUST00000176860.2 |
Bco1
|
beta-carotene oxygenase 1 |
| chr7_-_126992776 | 0.31 |
ENSMUST00000165495.2
ENSMUST00000106303.3 ENSMUST00000074249.7 |
E430018J23Rik
|
RIKEN cDNA E430018J23 gene |
| chr11_+_69011230 | 0.31 |
ENSMUST00000024543.3
|
Hes7
|
hes family bHLH transcription factor 7 |
| chrX_+_158623460 | 0.31 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_+_131568167 | 0.31 |
ENSMUST00000045840.5
|
Gpr26
|
G protein-coupled receptor 26 |
| chr8_-_85500998 | 0.30 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X |
| chr16_-_92494203 | 0.30 |
ENSMUST00000113956.10
|
Runx1
|
runt related transcription factor 1 |
| chr4_-_135221810 | 0.30 |
ENSMUST00000105856.9
|
Nipal3
|
NIPA-like domain containing 3 |
| chr19_+_8828132 | 0.30 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr5_+_115983292 | 0.30 |
ENSMUST00000137952.8
ENSMUST00000148245.8 |
Cit
|
citron |
| chr8_-_106052884 | 0.30 |
ENSMUST00000210412.2
ENSMUST00000210801.2 ENSMUST00000070508.8 |
Lrrc29
|
leucine rich repeat containing 29 |
| chr1_+_155911518 | 0.30 |
ENSMUST00000133152.2
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr19_+_3372296 | 0.29 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr16_+_43993599 | 0.29 |
ENSMUST00000119746.8
ENSMUST00000088356.10 ENSMUST00000169582.3 |
Usf3
|
upstream transcription factor family member 3 |
| chr9_+_38164070 | 0.29 |
ENSMUST00000213129.2
|
Olfr143
|
olfactory receptor 143 |
| chr9_+_54858066 | 0.29 |
ENSMUST00000034848.14
|
Psma4
|
proteasome subunit alpha 4 |
| chr2_-_18053595 | 0.29 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr7_-_103734672 | 0.29 |
ENSMUST00000057104.7
|
Olfr645
|
olfactory receptor 645 |
| chrX_+_135723420 | 0.29 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr16_+_14523696 | 0.29 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr2_-_69416365 | 0.29 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr9_-_32452885 | 0.28 |
ENSMUST00000016231.14
|
Fli1
|
Friend leukemia integration 1 |
| chr14_+_25459630 | 0.28 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr8_-_32408864 | 0.28 |
ENSMUST00000073884.7
ENSMUST00000238812.2 |
Nrg1
|
neuregulin 1 |
| chr9_-_87613301 | 0.28 |
ENSMUST00000034991.8
|
Tbx18
|
T-box18 |
| chr15_+_98390278 | 0.28 |
ENSMUST00000205772.3
ENSMUST00000216822.2 |
Olfr279
|
olfactory receptor 279 |
| chr1_-_63153414 | 0.28 |
ENSMUST00000153992.2
ENSMUST00000165066.8 ENSMUST00000172416.8 ENSMUST00000137511.8 |
Ino80d
|
INO80 complex subunit D |
| chr10_+_122514669 | 0.28 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr7_-_24831892 | 0.28 |
ENSMUST00000108418.11
ENSMUST00000175774.9 ENSMUST00000108415.10 ENSMUST00000098679.10 ENSMUST00000108417.10 ENSMUST00000108416.10 ENSMUST00000108413.8 ENSMUST00000176408.8 |
Pou2f2
|
POU domain, class 2, transcription factor 2 |
| chr11_+_70453806 | 0.27 |
ENSMUST00000079244.12
ENSMUST00000102558.11 |
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr1_+_159351337 | 0.27 |
ENSMUST00000192069.6
|
Tnr
|
tenascin R |
| chr8_+_85807566 | 0.27 |
ENSMUST00000140621.2
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chrX_+_100892981 | 0.27 |
ENSMUST00000124279.6
ENSMUST00000101339.8 |
Nhsl2
|
NHS-like 2 |
| chrX_+_158491589 | 0.27 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr9_-_110237276 | 0.27 |
ENSMUST00000040021.12
|
Ptpn23
|
protein tyrosine phosphatase, non-receptor type 23 |
| chr1_+_107456731 | 0.27 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr14_-_50564218 | 0.27 |
ENSMUST00000217152.2
|
Olfr734
|
olfactory receptor 734 |
| chr4_+_35152056 | 0.26 |
ENSMUST00000058595.7
|
Ifnk
|
interferon kappa |
| chr15_-_77480311 | 0.26 |
ENSMUST00000089465.6
|
Apol10b
|
apolipoprotein L 10B |
| chr14_-_50586329 | 0.26 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr11_+_115790768 | 0.26 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr18_+_61238714 | 0.26 |
ENSMUST00000237706.2
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr5_-_104125226 | 0.26 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_-_171061902 | 0.26 |
ENSMUST00000079957.12
|
Fcer1g
|
Fc receptor, IgE, high affinity I, gamma polypeptide |
| chr11_-_70545450 | 0.25 |
ENSMUST00000018437.3
|
Pfn1
|
profilin 1 |
| chr17_+_34362281 | 0.25 |
ENSMUST00000236519.2
|
H2-DMb2
|
histocompatibility 2, class II, locus Mb2 |
| chr18_+_37580692 | 0.25 |
ENSMUST00000052387.5
|
Pcdhb14
|
protocadherin beta 14 |
| chr14_+_28226697 | 0.25 |
ENSMUST00000063465.12
|
Wnt5a
|
wingless-type MMTV integration site family, member 5A |
| chr8_-_64659004 | 0.25 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr1_+_17672117 | 0.25 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chr9_-_106353571 | 0.24 |
ENSMUST00000123555.8
ENSMUST00000125850.2 |
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr10_-_120312374 | 0.24 |
ENSMUST00000072777.14
ENSMUST00000159699.2 |
Hmga2
|
high mobility group AT-hook 2 |
| chr9_+_44893077 | 0.24 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chr3_-_52012462 | 0.24 |
ENSMUST00000121440.4
|
Maml3
|
mastermind like transcriptional coactivator 3 |
| chr3_+_103767581 | 0.24 |
ENSMUST00000029433.9
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr15_+_82159398 | 0.24 |
ENSMUST00000023095.14
ENSMUST00000230365.2 |
Septin3
|
septin 3 |
| chr2_-_24365607 | 0.24 |
ENSMUST00000028355.11
|
Pax8
|
paired box 8 |
| chr8_+_109429732 | 0.24 |
ENSMUST00000188994.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr17_-_47143214 | 0.24 |
ENSMUST00000233537.2
|
Bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr2_-_45002902 | 0.24 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr4_-_66322695 | 0.24 |
ENSMUST00000084496.3
|
Astn2
|
astrotactin 2 |
| chr7_+_27879650 | 0.24 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr16_+_32090286 | 0.24 |
ENSMUST00000093183.5
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr16_-_32876544 | 0.24 |
ENSMUST00000115100.9
|
Iqcg
|
IQ motif containing G |
| chr14_+_54878409 | 0.24 |
ENSMUST00000169818.3
|
Gm17606
|
predicted gene, 17606 |
| chr4_-_123421441 | 0.24 |
ENSMUST00000147228.8
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr2_+_89804937 | 0.23 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chrM_+_3906 | 0.23 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr8_-_106723026 | 0.23 |
ENSMUST00000227363.2
ENSMUST00000081998.13 |
Dpep2
|
dipeptidase 2 |
| chr4_-_66322750 | 0.23 |
ENSMUST00000068214.11
|
Astn2
|
astrotactin 2 |
| chr10_-_129463803 | 0.23 |
ENSMUST00000204979.3
|
Olfr798
|
olfactory receptor 798 |
| chr11_-_97877219 | 0.23 |
ENSMUST00000107565.3
ENSMUST00000107564.2 ENSMUST00000017561.15 |
Plxdc1
|
plexin domain containing 1 |
| chr1_-_158785937 | 0.23 |
ENSMUST00000159861.9
|
Pappa2
|
pappalysin 2 |
| chr16_+_43184191 | 0.23 |
ENSMUST00000156367.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr14_+_51366306 | 0.23 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6 |
| chr3_-_89959739 | 0.23 |
ENSMUST00000199929.2
ENSMUST00000090908.11 ENSMUST00000198322.5 ENSMUST00000196843.5 |
Ubap2l
|
ubiquitin-associated protein 2-like |
| chr18_-_78185334 | 0.22 |
ENSMUST00000160639.2
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr4_+_136013372 | 0.22 |
ENSMUST00000069195.5
ENSMUST00000130658.2 |
Zfp46
|
zinc finger protein 46 |
| chr13_-_23650045 | 0.22 |
ENSMUST00000041674.14
ENSMUST00000110434.2 |
Btn1a1
|
butyrophilin, subfamily 1, member A1 |
| chr2_+_43445333 | 0.22 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr9_-_114325630 | 0.22 |
ENSMUST00000054414.5
|
Ccr4
|
chemokine (C-C motif) receptor 4 |
| chr2_-_167030706 | 0.22 |
ENSMUST00000207917.2
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr16_+_62635039 | 0.22 |
ENSMUST00000055557.6
|
Stx19
|
syntaxin 19 |
| chr1_-_78173744 | 0.22 |
ENSMUST00000087086.7
|
Pax3
|
paired box 3 |
| chr1_+_63655127 | 0.22 |
ENSMUST00000226288.2
|
Gm39653
|
predicted gene, 39653 |
| chr17_-_45125468 | 0.22 |
ENSMUST00000159943.8
ENSMUST00000160673.8 |
Runx2
|
runt related transcription factor 2 |
| chr14_+_54862762 | 0.22 |
ENSMUST00000097177.5
|
Psmb11
|
proteasome (prosome, macropain) subunit, beta type, 11 |
| chr2_-_111843053 | 0.22 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr10_+_127256993 | 0.22 |
ENSMUST00000170336.8
|
R3hdm2
|
R3H domain containing 2 |
| chr1_-_162687488 | 0.22 |
ENSMUST00000134098.8
ENSMUST00000111518.3 |
Fmo1
|
flavin containing monooxygenase 1 |
| chr9_+_38382403 | 0.22 |
ENSMUST00000214377.2
|
Olfr905
|
olfactory receptor 905 |
| chr19_-_34618135 | 0.21 |
ENSMUST00000087357.5
|
Ifit1bl2
|
interferon induced protein with tetratricopeptide repeats 1B like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf2_Ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.3 | 1.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 1.4 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.3 | 0.9 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.2 | 0.7 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.2 | 0.6 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.2 | 0.6 | GO:0021627 | olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.2 | 1.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 0.6 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.2 | 0.7 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 | 0.5 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.2 | 0.6 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.9 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 0.9 | GO:0035128 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 | 0.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 1.2 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.5 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.3 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.3 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.4 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.3 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.1 | 0.8 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.3 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 0.4 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.1 | 0.2 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.2 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.2 | GO:1904635 | glomerular visceral epithelial cell apoptotic process(GO:1903210) regulation of glomerular visceral epithelial cell apoptotic process(GO:1904633) positive regulation of glomerular visceral epithelial cell apoptotic process(GO:1904635) positive regulation of progesterone biosynthetic process(GO:2000184) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.1 | 0.4 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.1 | 0.2 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) |
| 0.1 | 0.2 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.1 | 0.2 | GO:0072233 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.3 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.1 | 0.2 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.2 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.1 | 2.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.2 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 | 0.2 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.9 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.2 | GO:1904799 | regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.1 | 0.3 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.2 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.2 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0072249 | metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.7 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0042701 | progesterone secretion(GO:0042701) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.5 | GO:0070572 | positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 1.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 1.0 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.3 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.7 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.0 | GO:0039519 | modulation by virus of host autophagy(GO:0039519) |
| 0.0 | 0.1 | GO:0072198 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) |
| 0.0 | 1.4 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 1.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.2 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.1 | GO:0061152 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0046722 | lactic acid secretion(GO:0046722) metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 | 0.1 | GO:1904092 | regulation of autophagic cell death(GO:1904092) negative regulation of autophagic cell death(GO:1904093) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.0 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0035037 | sperm entry(GO:0035037) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.1 | GO:0070781 | protein biotinylation(GO:0009305) response to biotin(GO:0070781) histone biotinylation(GO:0071110) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.4 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.0 | GO:1990791 | regulation of retinal ganglion cell axon guidance(GO:0090259) dorsal root ganglion development(GO:1990791) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.0 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.2 | GO:0072173 | metanephric tubule morphogenesis(GO:0072173) |
| 0.0 | 0.2 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.0 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.0 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 0.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.0 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.0 | GO:0097273 | creatinine homeostasis(GO:0097273) cellular ammonia homeostasis(GO:0097275) cellular creatinine homeostasis(GO:0097276) cellular urea homeostasis(GO:0097277) |
| 0.0 | 0.0 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.1 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.0 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.4 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.7 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.3 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.9 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.2 | GO:1990843 | subsarcolemmal mitochondrion(GO:1990843) interfibrillar mitochondrion(GO:1990844) |
| 0.1 | 0.2 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 1.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.5 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.0 | 0.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.5 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.7 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.4 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.4 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.9 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.9 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.5 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 2.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 2.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.6 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.6 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0004077 | biotin-[acetyl-CoA-carboxylase] ligase activity(GO:0004077) biotin-[methylcrotonoyl-CoA-carboxylase] ligase activity(GO:0004078) biotin-[methylmalonyl-CoA-carboxytransferase] ligase activity(GO:0004079) biotin-[propionyl-CoA-carboxylase (ATP-hydrolyzing)] ligase activity(GO:0004080) biotin-protein ligase activity(GO:0018271) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.0 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 2.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 3.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |