Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
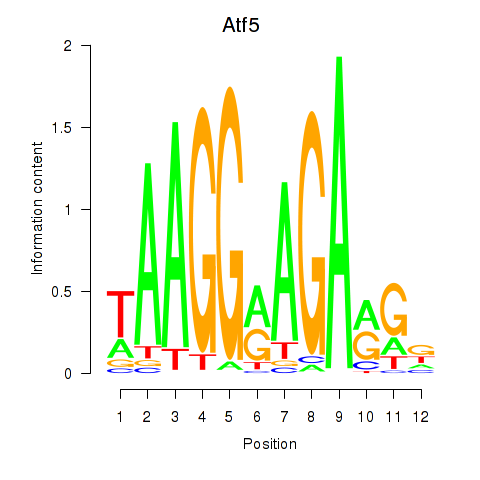
Results for Atf5
Z-value: 1.50
Transcription factors associated with Atf5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf5
|
ENSMUSG00000038539.16 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf5 | mm39_v1_chr7_-_44465043_44465082 | 0.74 | 1.5e-01 | Click! |
Activity profile of Atf5 motif
Sorted Z-values of Atf5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_30331839 | 2.01 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr8_-_78244412 | 1.41 |
ENSMUST00000210922.2
ENSMUST00000210519.2 |
Arhgap10
|
Rho GTPase activating protein 10 |
| chr10_-_85793639 | 1.24 |
ENSMUST00000001834.4
|
Rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr8_-_78244578 | 1.14 |
ENSMUST00000076316.6
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr6_-_16898440 | 1.06 |
ENSMUST00000031533.11
|
Tfec
|
transcription factor EC |
| chr11_+_101333238 | 0.90 |
ENSMUST00000107249.8
|
Rpl27
|
ribosomal protein L27 |
| chr6_+_116528102 | 0.89 |
ENSMUST00000122096.3
|
Eif4a3l2
|
eukaryotic translation initiation factor 4A3 like 2 |
| chr7_+_24069680 | 0.85 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr17_-_46342739 | 0.82 |
ENSMUST00000024747.14
|
Vegfa
|
vascular endothelial growth factor A |
| chr7_+_46496929 | 0.80 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chrX_-_17437801 | 0.74 |
ENSMUST00000177213.8
|
Fundc1
|
FUN14 domain containing 1 |
| chr17_+_85272459 | 0.71 |
ENSMUST00000234540.2
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr14_-_52252003 | 0.69 |
ENSMUST00000226522.2
|
Zfp219
|
zinc finger protein 219 |
| chr14_-_52341472 | 0.68 |
ENSMUST00000111610.12
ENSMUST00000164655.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr11_+_101333115 | 0.67 |
ENSMUST00000077856.13
|
Rpl27
|
ribosomal protein L27 |
| chr10_-_37014859 | 0.66 |
ENSMUST00000092584.6
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr15_-_77037972 | 0.62 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_+_41694317 | 0.61 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr18_-_43610829 | 0.60 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr11_-_61652866 | 0.59 |
ENSMUST00000004955.14
ENSMUST00000168115.8 |
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chrX_-_17438520 | 0.58 |
ENSMUST00000026016.13
|
Fundc1
|
FUN14 domain containing 1 |
| chr1_-_37575313 | 0.56 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr11_-_96714813 | 0.52 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr7_-_25089522 | 0.52 |
ENSMUST00000054301.14
|
Lipe
|
lipase, hormone sensitive |
| chr5_+_115479284 | 0.49 |
ENSMUST00000031508.5
|
Triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chrX_+_49930311 | 0.48 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr6_+_136305516 | 0.47 |
ENSMUST00000204966.2
ENSMUST00000077886.7 |
Eif4a3l1
|
eukaryotic translation initiation factor 4A3 like 1 |
| chr1_-_65218217 | 0.44 |
ENSMUST00000097709.11
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr18_+_35695339 | 0.44 |
ENSMUST00000237365.2
|
Matr3
|
matrin 3 |
| chr14_-_70391260 | 0.43 |
ENSMUST00000035612.7
|
Ccar2
|
cell cycle activator and apoptosis regulator 2 |
| chr3_+_89987749 | 0.42 |
ENSMUST00000127955.2
|
Tpm3
|
tropomyosin 3, gamma |
| chr5_+_100946493 | 0.42 |
ENSMUST00000016977.15
ENSMUST00000112898.8 ENSMUST00000112901.2 |
Mrps18c
|
mitochondrial ribosomal protein S18C |
| chr6_-_40562700 | 0.39 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr14_-_52341426 | 0.38 |
ENSMUST00000227536.2
ENSMUST00000227195.2 ENSMUST00000228815.2 ENSMUST00000228198.2 ENSMUST00000227458.2 ENSMUST00000228232.2 ENSMUST00000227242.2 ENSMUST00000228748.2 |
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr4_-_155845500 | 0.38 |
ENSMUST00000030903.12
|
Atad3a
|
ATPase family, AAA domain containing 3A |
| chr7_+_24310738 | 0.38 |
ENSMUST00000073325.6
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr12_+_4684620 | 0.37 |
ENSMUST00000217672.2
|
Itsn2
|
intersectin 2 |
| chr4_+_102427247 | 0.36 |
ENSMUST00000097950.9
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chrX_+_41239548 | 0.35 |
ENSMUST00000069619.14
|
Stag2
|
stromal antigen 2 |
| chr4_+_11758147 | 0.34 |
ENSMUST00000029871.12
ENSMUST00000108303.2 |
Cdh17
|
cadherin 17 |
| chr11_+_95733489 | 0.34 |
ENSMUST00000100532.10
ENSMUST00000036088.11 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chrX_-_74460168 | 0.33 |
ENSMUST00000033543.14
ENSMUST00000149863.3 ENSMUST00000114081.2 |
Cmc4
Mtcp1
|
C-x(9)-C motif containing 4 mature T cell proliferation 1 |
| chr4_-_133599616 | 0.33 |
ENSMUST00000157067.9
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr19_-_7460550 | 0.32 |
ENSMUST00000088169.7
|
Rtn3
|
reticulon 3 |
| chr9_+_45818250 | 0.32 |
ENSMUST00000216672.2
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr13_+_104424359 | 0.31 |
ENSMUST00000065766.7
|
Adamts6
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 6 |
| chr17_+_35439810 | 0.31 |
ENSMUST00000118793.2
|
Gm16181
|
predicted gene 16181 |
| chr19_+_55240357 | 0.29 |
ENSMUST00000225551.2
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr18_+_69633741 | 0.28 |
ENSMUST00000207214.2
ENSMUST00000201094.4 ENSMUST00000200703.4 ENSMUST00000202765.4 |
Tcf4
|
transcription factor 4 |
| chr16_-_10265204 | 0.27 |
ENSMUST00000051118.7
|
Tvp23a
|
trans-golgi network vesicle protein 23A |
| chr14_-_52252432 | 0.27 |
ENSMUST00000226527.2
|
Zfp219
|
zinc finger protein 219 |
| chr3_+_145281941 | 0.27 |
ENSMUST00000199033.5
ENSMUST00000098534.9 ENSMUST00000200574.5 ENSMUST00000196413.5 ENSMUST00000197604.3 |
Znhit6
|
zinc finger, HIT type 6 |
| chr11_+_4852212 | 0.26 |
ENSMUST00000142543.3
|
Thoc5
|
THO complex 5 |
| chr4_+_11579648 | 0.23 |
ENSMUST00000180239.2
|
Fsbp
|
fibrinogen silencer binding protein |
| chr2_-_120370333 | 0.23 |
ENSMUST00000171215.8
|
Zfp106
|
zinc finger protein 106 |
| chr19_-_7460600 | 0.22 |
ENSMUST00000235593.2
ENSMUST00000088171.12 ENSMUST00000065304.13 ENSMUST00000025667.14 |
Rtn3
|
reticulon 3 |
| chr15_-_77037756 | 0.22 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_-_59266511 | 0.20 |
ENSMUST00000043204.8
|
Fbxo33
|
F-box protein 33 |
| chr14_+_4198667 | 0.19 |
ENSMUST00000079419.12
ENSMUST00000100799.9 |
Rpl15
|
ribosomal protein L15 |
| chr4_-_141602190 | 0.19 |
ENSMUST00000036854.4
|
Efhd2
|
EF hand domain containing 2 |
| chr18_+_35695199 | 0.19 |
ENSMUST00000236860.2
ENSMUST00000166793.10 ENSMUST00000237780.2 ENSMUST00000236507.2 ENSMUST00000235960.2 ENSMUST00000237061.2 |
Matr3
|
matrin 3 |
| chr16_+_64672334 | 0.19 |
ENSMUST00000067744.8
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr1_-_53824307 | 0.18 |
ENSMUST00000185920.2
|
Stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr6_+_65567373 | 0.18 |
ENSMUST00000114236.2
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chrX_-_74460137 | 0.18 |
ENSMUST00000033542.11
|
Mtcp1
|
mature T cell proliferation 1 |
| chr14_-_52252318 | 0.18 |
ENSMUST00000228051.2
|
Zfp219
|
zinc finger protein 219 |
| chr11_-_101442663 | 0.18 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr2_-_153286361 | 0.16 |
ENSMUST00000109784.2
|
Nol4l
|
nucleolar protein 4-like |
| chr5_-_100946334 | 0.16 |
ENSMUST00000133845.8
|
Helq
|
helicase, POLQ-like |
| chr15_-_77037926 | 0.16 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_+_172235820 | 0.15 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr4_+_21727695 | 0.15 |
ENSMUST00000065928.11
ENSMUST00000102997.8 ENSMUST00000120679.8 |
Ccnc
|
cyclin C |
| chr10_+_108167973 | 0.15 |
ENSMUST00000095313.5
|
Pawr
|
PRKC, apoptosis, WT1, regulator |
| chr11_+_95733109 | 0.14 |
ENSMUST00000107714.9
ENSMUST00000107711.8 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr5_-_100946434 | 0.12 |
ENSMUST00000044684.14
|
Helq
|
helicase, POLQ-like |
| chr9_+_32027335 | 0.12 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr9_-_21042521 | 0.11 |
ENSMUST00000216874.2
ENSMUST00000214454.2 ENSMUST00000214864.2 |
Tyk2
|
tyrosine kinase 2 |
| chr3_+_99161070 | 0.11 |
ENSMUST00000029462.10
|
Tbx15
|
T-box 15 |
| chr18_+_35695485 | 0.11 |
ENSMUST00000235199.2
ENSMUST00000237744.2 ENSMUST00000236276.2 |
Matr3
|
matrin 3 |
| chr2_+_26800757 | 0.11 |
ENSMUST00000102898.5
|
Rpl7a
|
ribosomal protein L7A |
| chr1_+_59555423 | 0.10 |
ENSMUST00000114243.8
|
Gm973
|
predicted gene 973 |
| chr6_+_129326927 | 0.10 |
ENSMUST00000065289.6
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr19_-_18978463 | 0.09 |
ENSMUST00000040153.15
ENSMUST00000112828.8 |
Rorb
|
RAR-related orphan receptor beta |
| chr19_+_27194757 | 0.09 |
ENSMUST00000047645.13
ENSMUST00000167487.8 |
Vldlr
|
very low density lipoprotein receptor |
| chr14_-_10185523 | 0.08 |
ENSMUST00000223702.2
ENSMUST00000223762.2 ENSMUST00000112669.10 ENSMUST00000163392.3 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chr4_+_54947976 | 0.08 |
ENSMUST00000098070.10
|
Zfp462
|
zinc finger protein 462 |
| chr4_+_21727727 | 0.08 |
ENSMUST00000108240.3
|
Ccnc
|
cyclin C |
| chr5_+_137759934 | 0.08 |
ENSMUST00000110983.3
ENSMUST00000031738.5 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr7_+_126184141 | 0.07 |
ENSMUST00000137646.8
|
Apobr
|
apolipoprotein B receptor |
| chr11_-_95733235 | 0.06 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr2_+_10377237 | 0.06 |
ENSMUST00000114864.9
ENSMUST00000116594.9 ENSMUST00000041105.7 |
Sfmbt2
|
Scm-like with four mbt domains 2 |
| chr5_+_20112500 | 0.06 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr3_+_89986925 | 0.06 |
ENSMUST00000118566.8
ENSMUST00000119158.8 |
Tpm3
|
tropomyosin 3, gamma |
| chrX_+_40490353 | 0.05 |
ENSMUST00000165288.2
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr16_-_45830575 | 0.05 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr10_+_89906956 | 0.05 |
ENSMUST00000183109.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_-_17438470 | 0.05 |
ENSMUST00000176638.8
|
Fundc1
|
FUN14 domain containing 1 |
| chr8_-_106074585 | 0.04 |
ENSMUST00000014922.5
|
Fhod1
|
formin homology 2 domain containing 1 |
| chrX_+_55939732 | 0.04 |
ENSMUST00000136396.8
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr8_+_82499732 | 0.03 |
ENSMUST00000170160.8
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr7_+_51271742 | 0.03 |
ENSMUST00000032710.7
|
Slc17a6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 |
| chr15_-_77129706 | 0.03 |
ENSMUST00000228361.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_+_27194411 | 0.03 |
ENSMUST00000164746.8
ENSMUST00000172302.8 |
Vldlr
|
very low density lipoprotein receptor |
| chr15_-_77129786 | 0.03 |
ENSMUST00000228558.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr11_+_49231361 | 0.03 |
ENSMUST00000062719.2
|
Olfr1390
|
olfactory receptor 1390 |
| chr1_-_157084252 | 0.03 |
ENSMUST00000134543.8
|
Rasal2
|
RAS protein activator like 2 |
| chr14_-_10185481 | 0.03 |
ENSMUST00000225640.2
ENSMUST00000225294.2 ENSMUST00000224389.2 ENSMUST00000223927.2 |
3830406C13Rik
|
RIKEN cDNA 3830406C13 gene |
| chr2_+_177783713 | 0.03 |
ENSMUST00000103066.10
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr17_+_20639425 | 0.03 |
ENSMUST00000170076.2
|
Vmn1r224
|
vomeronasal 1 receptor 224 |
| chrX_+_74460275 | 0.03 |
ENSMUST00000118428.8
ENSMUST00000114074.8 ENSMUST00000133781.8 |
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr8_+_106075475 | 0.03 |
ENSMUST00000073149.7
|
Slc9a5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5 |
| chr2_-_86938729 | 0.03 |
ENSMUST00000099862.2
|
Olfr259
|
olfactory receptor 259 |
| chr1_+_153541412 | 0.03 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr14_+_71127882 | 0.02 |
ENSMUST00000227697.2
|
Gfra2
|
glial cell line derived neurotrophic factor family receptor alpha 2 |
| chr10_+_129186434 | 0.02 |
ENSMUST00000077024.2
|
Olfr782
|
olfactory receptor 782 |
| chr2_-_86254018 | 0.02 |
ENSMUST00000105213.5
|
Olfr1062
|
olfactory receptor 1062 |
| chr7_-_4999042 | 0.02 |
ENSMUST00000162502.2
|
Zfp579
|
zinc finger protein 579 |
| chr5_+_20112704 | 0.02 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr8_+_55407872 | 0.02 |
ENSMUST00000033915.9
|
Gpm6a
|
glycoprotein m6a |
| chr12_+_89779237 | 0.02 |
ENSMUST00000110133.9
ENSMUST00000110130.4 |
Nrxn3
|
neurexin III |
| chr5_-_147244074 | 0.02 |
ENSMUST00000031650.4
|
Cdx2
|
caudal type homeobox 2 |
| chr11_+_49414548 | 0.02 |
ENSMUST00000077143.7
|
Olfr1383
|
olfactory receptor 1383 |
| chr12_+_89779178 | 0.02 |
ENSMUST00000238943.2
|
Nrxn3
|
neurexin III |
| chr4_+_118750282 | 0.02 |
ENSMUST00000094830.2
|
Olfr1330
|
olfactory receptor 1330 |
| chr14_+_122712809 | 0.02 |
ENSMUST00000075888.6
|
Zic2
|
zinc finger protein of the cerebellum 2 |
| chr2_+_162829250 | 0.01 |
ENSMUST00000018012.14
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr16_+_84312009 | 0.01 |
ENSMUST00000209558.2
|
A730009L09Rik
|
RIKEN cDNA A730009L09 gene |
| chr17_+_88228405 | 0.01 |
ENSMUST00000179601.2
|
Rpl36-ps4
|
ribosomal protein L36, pseudogene 4 |
| chr16_-_26810402 | 0.01 |
ENSMUST00000231299.2
|
Gmnc
|
geminin coiled-coil domain containing |
| chr16_+_19305543 | 0.01 |
ENSMUST00000074739.4
|
Olfr166
|
olfactory receptor 166 |
| chr4_-_91264670 | 0.01 |
ENSMUST00000107109.9
ENSMUST00000107111.9 ENSMUST00000107120.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr1_+_153541339 | 0.01 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr16_+_17716480 | 0.01 |
ENSMUST00000055374.8
|
Tssk2
|
testis-specific serine kinase 2 |
| chr10_-_20600442 | 0.01 |
ENSMUST00000170265.8
|
Pde7b
|
phosphodiesterase 7B |
| chr7_-_105249308 | 0.01 |
ENSMUST00000210531.2
ENSMUST00000033185.10 |
Hpx
|
hemopexin |
| chr2_+_111158766 | 0.01 |
ENSMUST00000090326.2
|
Olfr1281
|
olfactory receptor 1281 |
| chr3_-_93846078 | 0.01 |
ENSMUST00000177735.2
|
Tdpoz9-ps1
|
TD and POZ domain containing 9, pseudogene 1 |
| chr17_+_72076728 | 0.01 |
ENSMUST00000230305.2
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr15_+_22549108 | 0.01 |
ENSMUST00000163361.8
|
Cdh18
|
cadherin 18 |
| chr17_-_38393890 | 0.01 |
ENSMUST00000059560.5
|
Olfr131
|
olfactory receptor 131 |
| chr11_-_49004584 | 0.01 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr4_+_118516149 | 0.01 |
ENSMUST00000213189.3
|
Olfr62
|
olfactory receptor 62 |
| chr3_-_92366022 | 0.01 |
ENSMUST00000058142.4
|
Sprr3
|
small proline-rich protein 3 |
| chr5_+_119808890 | 0.01 |
ENSMUST00000121021.8
|
Tbx3
|
T-box 3 |
| chr10_-_78554104 | 0.01 |
ENSMUST00000005488.9
|
Casp14
|
caspase 14 |
| chr12_-_64521464 | 0.01 |
ENSMUST00000059833.8
|
Fscb
|
fibrous sheath CABYR binding protein |
| chr18_+_44467133 | 0.01 |
ENSMUST00000025349.12
ENSMUST00000115498.2 |
Myot
|
myotilin |
| chr3_+_113824181 | 0.01 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr7_+_102597674 | 0.01 |
ENSMUST00000052997.6
ENSMUST00000211329.2 |
Olfr574
|
olfactory receptor 574 |
| chr2_+_89845235 | 0.01 |
ENSMUST00000111507.4
|
Olfr1263
|
olfactory receptor 1263 |
| chr15_-_79048674 | 0.01 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chrX_-_166907286 | 0.00 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr16_+_26281885 | 0.00 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chr8_-_110765983 | 0.00 |
ENSMUST00000109222.4
|
Chst4
|
carbohydrate sulfotransferase 4 |
| chr2_+_36311412 | 0.00 |
ENSMUST00000071437.2
|
Olfr339
|
olfactory receptor 339 |
| chr14_+_55173936 | 0.00 |
ENSMUST00000227441.2
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr5_+_119809076 | 0.00 |
ENSMUST00000018748.9
|
Tbx3
|
T-box 3 |
| chr13_-_116446166 | 0.00 |
ENSMUST00000036060.13
|
Isl1
|
ISL1 transcription factor, LIM/homeodomain |
| chr10_+_23796254 | 0.00 |
ENSMUST00000051532.5
|
Taar1
|
trace amine-associated receptor 1 |
| chrX_+_37861548 | 0.00 |
ENSMUST00000050744.6
|
6030498E09Rik
|
RIKEN cDNA 6030498E09 gene |
| chr1_-_69726384 | 0.00 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr7_+_140072530 | 0.00 |
ENSMUST00000074897.5
|
Olfr535
|
olfactory receptor 535 |
| chr17_+_72076678 | 0.00 |
ENSMUST00000230427.2
ENSMUST00000229952.2 ENSMUST00000230333.2 |
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr2_-_36370140 | 0.00 |
ENSMUST00000100154.2
|
Olfr341
|
olfactory receptor 341 |
| chr7_-_106526664 | 0.00 |
ENSMUST00000072368.3
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr18_-_23174698 | 0.00 |
ENSMUST00000097651.10
|
Nol4
|
nucleolar protein 4 |
| chr5_-_86345729 | 0.00 |
ENSMUST00000031172.9
ENSMUST00000113372.2 |
Gnrhr
|
gonadotropin releasing hormone receptor |
| chr18_+_65715351 | 0.00 |
ENSMUST00000235669.2
ENSMUST00000183319.8 ENSMUST00000236523.2 ENSMUST00000237886.2 |
Zfp532
|
zinc finger protein 532 |
| chr7_-_37470184 | 0.00 |
ENSMUST00000176680.8
|
Zfp536
|
zinc finger protein 536 |
| chr14_+_55173696 | 0.00 |
ENSMUST00000037814.8
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr2_-_86048052 | 0.00 |
ENSMUST00000099902.2
|
Olfr1046
|
olfactory receptor 1046 |
| chr14_-_124914516 | 0.00 |
ENSMUST00000095529.10
|
Fgf14
|
fibroblast growth factor 14 |
| chr1_-_65112680 | 0.00 |
ENSMUST00000114064.3
|
Crygc
|
crystallin, gamma C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.4 | 1.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.3 | 0.8 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.1 | 0.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.4 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.7 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 1.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.8 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 0.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 | 0.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0060450 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) positive regulation of hindgut contraction(GO:0060450) regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 0.0 | 0.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.4 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.4 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 1.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.4 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.7 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 2.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:1904949 | ATPase complex(GO:1904949) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.3 | 0.8 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 2.6 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |