Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
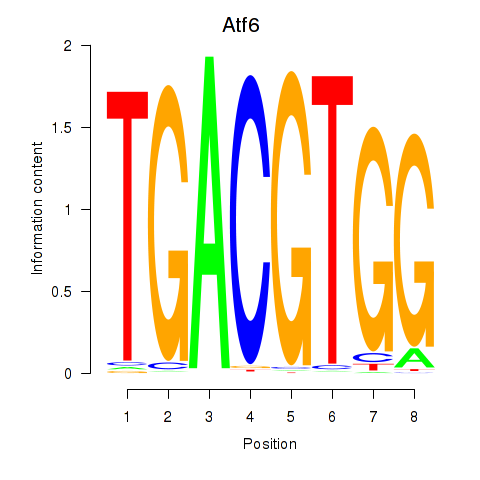
Results for Atf6
Z-value: 1.69
Transcription factors associated with Atf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf6
|
ENSMUSG00000026663.7 | activating transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf6 | mm39_v1_chr1_-_170695328_170695354 | -0.77 | 1.2e-01 | Click! |
Activity profile of Atf6 motif
Sorted Z-values of Atf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_80145805 | 1.03 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr11_-_69812016 | 1.01 |
ENSMUST00000108607.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chrX_-_133709733 | 0.98 |
ENSMUST00000035559.11
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr6_+_83092476 | 0.97 |
ENSMUST00000032114.8
|
Mogs
|
mannosyl-oligosaccharide glucosidase |
| chr3_+_104545974 | 0.74 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr4_-_149858694 | 0.73 |
ENSMUST00000105686.3
|
Slc25a33
|
solute carrier family 25, member 33 |
| chr6_+_88061464 | 0.60 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr6_-_124441731 | 0.58 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr19_-_41836514 | 0.57 |
ENSMUST00000059231.4
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr8_+_13388745 | 0.55 |
ENSMUST00000209885.2
ENSMUST00000209396.2 |
Tfdp1
|
transcription factor Dp 1 |
| chr4_-_43000450 | 0.54 |
ENSMUST00000030164.8
|
Vcp
|
valosin containing protein |
| chr7_-_45116197 | 0.54 |
ENSMUST00000211195.2
ENSMUST00000210019.2 |
Bax
|
BCL2-associated X protein |
| chr14_+_32507920 | 0.54 |
ENSMUST00000039191.8
ENSMUST00000227060.2 ENSMUST00000228481.2 |
Tmem273
|
transmembrane protein 273 |
| chr7_-_113853894 | 0.53 |
ENSMUST00000033012.9
|
Copb1
|
coatomer protein complex, subunit beta 1 |
| chr17_-_36353582 | 0.52 |
ENSMUST00000058801.15
ENSMUST00000080015.12 ENSMUST00000077960.7 |
H2-T22
|
histocompatibility 2, T region locus 22 |
| chr1_-_93373145 | 0.51 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr7_-_45116216 | 0.50 |
ENSMUST00000210392.2
ENSMUST00000211365.2 |
Bax
|
BCL2-associated X protein |
| chr15_-_98626002 | 0.50 |
ENSMUST00000003445.8
|
Fkbp11
|
FK506 binding protein 11 |
| chr1_-_93373364 | 0.49 |
ENSMUST00000190321.7
ENSMUST00000042498.14 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr3_+_89366425 | 0.49 |
ENSMUST00000029564.12
|
Pmvk
|
phosphomevalonate kinase |
| chr7_-_45116316 | 0.48 |
ENSMUST00000033093.10
|
Bax
|
BCL2-associated X protein |
| chr2_-_105229653 | 0.48 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr16_-_94171853 | 0.46 |
ENSMUST00000113914.8
ENSMUST00000113905.8 |
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr5_+_45677571 | 0.45 |
ENSMUST00000156481.8
ENSMUST00000119579.3 ENSMUST00000118833.3 |
Med28
|
mediator complex subunit 28 |
| chr9_+_44290832 | 0.45 |
ENSMUST00000161318.8
ENSMUST00000217019.2 ENSMUST00000160902.8 |
Hyou1
|
hypoxia up-regulated 1 |
| chr6_+_88442391 | 0.44 |
ENSMUST00000032165.16
|
Ruvbl1
|
RuvB-like protein 1 |
| chr10_+_88566918 | 0.44 |
ENSMUST00000116234.9
|
Arl1
|
ADP-ribosylation factor-like 1 |
| chr15_+_44291470 | 0.43 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr5_-_137531463 | 0.42 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr13_-_111626562 | 0.42 |
ENSMUST00000091236.11
ENSMUST00000047627.14 |
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr7_+_65759198 | 0.41 |
ENSMUST00000036372.8
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr1_-_131025562 | 0.41 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr13_-_119545479 | 0.41 |
ENSMUST00000223268.2
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr11_-_69811717 | 0.40 |
ENSMUST00000152589.2
ENSMUST00000108612.8 ENSMUST00000108611.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr7_-_30814652 | 0.40 |
ENSMUST00000168884.8
ENSMUST00000108102.9 |
Hpn
|
hepsin |
| chr4_+_102843540 | 0.39 |
ENSMUST00000030248.12
ENSMUST00000125417.9 ENSMUST00000169211.3 |
Dynlt5
|
dynein light chain Tctex-type 5 |
| chr4_+_47474652 | 0.39 |
ENSMUST00000065678.6
|
Sec61b
|
Sec61 beta subunit |
| chr6_+_134617903 | 0.39 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr12_-_13299197 | 0.39 |
ENSMUST00000071103.10
|
Ddx1
|
DEAD box helicase 1 |
| chr11_-_93776580 | 0.38 |
ENSMUST00000066888.10
|
Utp18
|
UTP18 small subunit processome component |
| chr5_-_52628825 | 0.38 |
ENSMUST00000198008.5
ENSMUST00000059428.7 |
Ccdc149
|
coiled-coil domain containing 149 |
| chr17_+_35117905 | 0.38 |
ENSMUST00000097342.10
ENSMUST00000013931.12 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr5_-_137531413 | 0.38 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr5_+_30824121 | 0.38 |
ENSMUST00000144742.6
ENSMUST00000149759.2 ENSMUST00000199320.5 |
Cenpa
|
centromere protein A |
| chr12_-_13299136 | 0.38 |
ENSMUST00000221623.2
|
Ddx1
|
DEAD box helicase 1 |
| chr19_+_23118545 | 0.37 |
ENSMUST00000036884.3
|
Klf9
|
Kruppel-like factor 9 |
| chrX_+_133618693 | 0.37 |
ENSMUST00000113201.8
ENSMUST00000051256.10 ENSMUST00000113199.8 ENSMUST00000035748.14 ENSMUST00000113198.8 ENSMUST00000113197.2 |
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr11_-_69872050 | 0.37 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr9_+_44290787 | 0.37 |
ENSMUST00000066601.13
|
Hyou1
|
hypoxia up-regulated 1 |
| chr13_-_119545520 | 0.36 |
ENSMUST00000069902.13
ENSMUST00000099149.10 ENSMUST00000109204.8 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr16_+_17577464 | 0.36 |
ENSMUST00000129199.8
|
Klhl22
|
kelch-like 22 |
| chr1_+_157334298 | 0.36 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr1_+_157334347 | 0.36 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr16_+_17577493 | 0.36 |
ENSMUST00000165790.9
|
Klhl22
|
kelch-like 22 |
| chr10_+_128158413 | 0.36 |
ENSMUST00000219836.2
|
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr12_+_84363603 | 0.35 |
ENSMUST00000045931.12
|
Zfp410
|
zinc finger protein 410 |
| chr17_-_29566774 | 0.35 |
ENSMUST00000095427.12
ENSMUST00000118366.9 |
Mtch1
|
mitochondrial carrier 1 |
| chr11_-_69811890 | 0.35 |
ENSMUST00000108609.8
ENSMUST00000108608.8 ENSMUST00000164359.8 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr15_-_79571977 | 0.35 |
ENSMUST00000023061.7
|
Josd1
|
Josephin domain containing 1 |
| chr8_+_26210064 | 0.35 |
ENSMUST00000068916.16
ENSMUST00000139836.8 |
Plpp5
|
phospholipid phosphatase 5 |
| chr6_-_115735935 | 0.35 |
ENSMUST00000072933.13
|
Tmem40
|
transmembrane protein 40 |
| chr13_+_93440265 | 0.35 |
ENSMUST00000109494.8
|
Homer1
|
homer scaffolding protein 1 |
| chrX_-_135642025 | 0.34 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr4_+_47474715 | 0.34 |
ENSMUST00000137461.8
ENSMUST00000125622.2 |
Sec61b
|
Sec61 beta subunit |
| chr17_+_29709723 | 0.34 |
ENSMUST00000024811.9
|
Pim1
|
proviral integration site 1 |
| chr6_-_131270136 | 0.34 |
ENSMUST00000032307.12
|
Magohb
|
mago homolog B, exon junction complex core component |
| chrX_+_49930311 | 0.34 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr3_-_143910926 | 0.33 |
ENSMUST00000120539.8
ENSMUST00000196264.5 |
Lmo4
|
LIM domain only 4 |
| chr11_+_87938626 | 0.33 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr9_+_120321557 | 0.33 |
ENSMUST00000007139.6
|
Eif1b
|
eukaryotic translation initiation factor 1B |
| chr8_-_86281946 | 0.33 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr4_+_46489248 | 0.33 |
ENSMUST00000030018.5
|
Nans
|
N-acetylneuraminic acid synthase (sialic acid synthase) |
| chr4_-_150994083 | 0.33 |
ENSMUST00000105674.8
ENSMUST00000105673.8 |
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr13_+_93440572 | 0.33 |
ENSMUST00000109493.9
|
Homer1
|
homer scaffolding protein 1 |
| chr10_-_84369831 | 0.33 |
ENSMUST00000167671.2
ENSMUST00000053871.5 |
Ckap4
|
cytoskeleton-associated protein 4 |
| chr7_-_84339045 | 0.32 |
ENSMUST00000209165.2
|
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr15_-_44291699 | 0.32 |
ENSMUST00000038719.8
|
Nudcd1
|
NudC domain containing 1 |
| chr2_+_75489596 | 0.32 |
ENSMUST00000111964.8
ENSMUST00000111962.8 ENSMUST00000111961.8 ENSMUST00000164947.9 ENSMUST00000090792.11 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr3_+_123061094 | 0.32 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr15_+_78915071 | 0.32 |
ENSMUST00000006544.9
ENSMUST00000171999.9 |
Gcat
Gcat
|
glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) |
| chr3_+_87754057 | 0.32 |
ENSMUST00000107581.9
|
Sh2d2a
|
SH2 domain containing 2A |
| chr19_-_41252370 | 0.31 |
ENSMUST00000237871.2
ENSMUST00000025989.10 |
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr11_-_72106418 | 0.31 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr2_+_164587948 | 0.31 |
ENSMUST00000109327.4
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr11_+_96209093 | 0.31 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr4_+_33031342 | 0.31 |
ENSMUST00000124992.8
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr3_+_131270529 | 0.30 |
ENSMUST00000029666.14
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr11_-_6217718 | 0.30 |
ENSMUST00000004507.11
ENSMUST00000151446.2 |
Ddx56
|
DEAD box helicase 56 |
| chr16_-_94171533 | 0.30 |
ENSMUST00000113910.8
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr17_+_35117438 | 0.30 |
ENSMUST00000114033.9
ENSMUST00000078061.13 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr9_+_108216433 | 0.30 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr6_+_116314975 | 0.30 |
ENSMUST00000079012.13
ENSMUST00000101032.10 ENSMUST00000123405.8 ENSMUST00000204657.3 ENSMUST00000203116.2 ENSMUST00000203193.3 ENSMUST00000126376.8 |
Marchf8
|
membrane associated ring-CH-type finger 8 |
| chr11_+_94520567 | 0.30 |
ENSMUST00000021239.7
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr15_-_98729333 | 0.29 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr9_-_62029877 | 0.29 |
ENSMUST00000185675.7
|
Glce
|
glucuronyl C5-epimerase |
| chr6_-_119825081 | 0.29 |
ENSMUST00000183703.8
ENSMUST00000183911.8 |
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr5_-_124490296 | 0.29 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr8_+_26210484 | 0.29 |
ENSMUST00000210629.2
|
Plpp5
|
phospholipid phosphatase 5 |
| chr5_-_65650269 | 0.29 |
ENSMUST00000121661.8
|
Smim14
|
small integral membrane protein 14 |
| chr11_-_103588605 | 0.29 |
ENSMUST00000021329.14
|
Gosr2
|
golgi SNAP receptor complex member 2 |
| chr19_-_53026965 | 0.28 |
ENSMUST00000183274.8
ENSMUST00000182097.2 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr8_-_106692668 | 0.28 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chrX_-_135641869 | 0.28 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr19_+_8849000 | 0.28 |
ENSMUST00000096255.7
|
Ubxn1
|
UBX domain protein 1 |
| chr13_+_33187205 | 0.28 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr9_+_107468146 | 0.28 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr9_+_57428490 | 0.27 |
ENSMUST00000000090.8
|
Cox5a
|
cytochrome c oxidase subunit 5A |
| chr12_-_84455764 | 0.27 |
ENSMUST00000120942.8
ENSMUST00000110272.9 |
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr17_-_34250616 | 0.27 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr12_+_51424343 | 0.27 |
ENSMUST00000219434.2
ENSMUST00000021335.7 |
Scfd1
|
Sec1 family domain containing 1 |
| chr8_-_13155431 | 0.27 |
ENSMUST00000164416.8
|
Pcid2
|
PCI domain containing 2 |
| chr12_-_59058780 | 0.27 |
ENSMUST00000021375.12
|
Sec23a
|
SEC23 homolog A, COPII coat complex component |
| chr1_-_161078723 | 0.27 |
ENSMUST00000051925.5
ENSMUST00000071718.12 |
Prdx6
|
peroxiredoxin 6 |
| chrX_-_133501874 | 0.26 |
ENSMUST00000033621.8
|
Gla
|
galactosidase, alpha |
| chr17_-_65920481 | 0.26 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr1_+_171173252 | 0.26 |
ENSMUST00000006579.5
|
Pfdn2
|
prefoldin 2 |
| chr2_+_121279842 | 0.26 |
ENSMUST00000110615.8
ENSMUST00000099475.12 |
Serf2
|
small EDRK-rich factor 2 |
| chr1_+_91468266 | 0.26 |
ENSMUST00000086843.11
|
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr11_-_102771751 | 0.26 |
ENSMUST00000021306.14
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr13_-_100969823 | 0.26 |
ENSMUST00000225922.2
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr19_+_8944369 | 0.25 |
ENSMUST00000052248.8
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr4_-_129271909 | 0.25 |
ENSMUST00000030610.3
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chr9_+_59198829 | 0.25 |
ENSMUST00000217570.2
ENSMUST00000026266.9 |
Adpgk
|
ADP-dependent glucokinase |
| chr19_-_46950355 | 0.25 |
ENSMUST00000236501.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr17_-_67661382 | 0.25 |
ENSMUST00000223982.2
ENSMUST00000224091.2 |
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr3_+_89366632 | 0.25 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr2_+_18677195 | 0.25 |
ENSMUST00000171845.8
ENSMUST00000061158.5 |
Commd3
|
COMM domain containing 3 |
| chr9_-_89996712 | 0.25 |
ENSMUST00000191353.2
ENSMUST00000085248.12 |
Morf4l1
|
mortality factor 4 like 1 |
| chr7_+_45522551 | 0.25 |
ENSMUST00000211234.2
|
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr10_-_18619658 | 0.24 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr4_+_88012836 | 0.24 |
ENSMUST00000097992.10
|
Focad
|
focadhesin |
| chr12_+_84455813 | 0.24 |
ENSMUST00000081828.13
|
Bbof1
|
basal body orientation factor 1 |
| chr12_+_72488625 | 0.24 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr17_+_25352353 | 0.24 |
ENSMUST00000162862.3
ENSMUST00000040729.9 |
Clcn7
|
chloride channel, voltage-sensitive 7 |
| chr3_+_40904253 | 0.24 |
ENSMUST00000048490.13
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr6_+_17694003 | 0.24 |
ENSMUST00000052113.12
ENSMUST00000081635.13 |
St7
|
suppression of tumorigenicity 7 |
| chr3_-_37778470 | 0.24 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr4_+_3938881 | 0.24 |
ENSMUST00000108386.8
ENSMUST00000121110.8 ENSMUST00000149544.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chrX_+_133501928 | 0.24 |
ENSMUST00000074950.11
ENSMUST00000113203.2 ENSMUST00000113202.8 ENSMUST00000050331.13 ENSMUST00000059297.6 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr5_+_143389573 | 0.23 |
ENSMUST00000110731.4
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr18_+_34994253 | 0.23 |
ENSMUST00000165033.2
|
Egr1
|
early growth response 1 |
| chr9_+_59446823 | 0.23 |
ENSMUST00000026262.8
|
Hexa
|
hexosaminidase A |
| chr11_-_102771806 | 0.23 |
ENSMUST00000107060.8
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr8_+_13807652 | 0.23 |
ENSMUST00000130173.9
ENSMUST00000043962.9 ENSMUST00000134645.8 |
Cdc16
|
CDC16 cell division cycle 16 |
| chr7_-_113875261 | 0.23 |
ENSMUST00000135570.8
|
Psma1
|
proteasome subunit alpha 1 |
| chr6_+_134012602 | 0.23 |
ENSMUST00000081028.13
ENSMUST00000111963.8 |
Etv6
|
ets variant 6 |
| chr19_+_8718837 | 0.23 |
ENSMUST00000177373.8
ENSMUST00000010254.16 |
Stx5a
|
syntaxin 5A |
| chr11_-_69812053 | 0.23 |
ENSMUST00000108613.10
ENSMUST00000043419.10 ENSMUST00000070996.11 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr14_+_70314727 | 0.23 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr7_-_44778050 | 0.23 |
ENSMUST00000209711.2
ENSMUST00000211037.2 ENSMUST00000209927.2 ENSMUST00000209815.2 ENSMUST00000210918.2 ENSMUST00000150350.9 |
Rpl13a
|
ribosomal protein L13A |
| chr5_-_123127346 | 0.22 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr17_-_25459086 | 0.22 |
ENSMUST00000038973.7
ENSMUST00000115154.11 |
Gnptg
|
N-acetylglucosamine-1-phosphotransferase, gamma subunit |
| chr11_+_62442502 | 0.22 |
ENSMUST00000136938.2
|
Ubb
|
ubiquitin B |
| chr19_+_8719033 | 0.22 |
ENSMUST00000176314.8
ENSMUST00000073430.14 ENSMUST00000175901.8 |
Stx5a
|
syntaxin 5A |
| chr13_-_100969878 | 0.22 |
ENSMUST00000067246.6
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr4_+_8535604 | 0.22 |
ENSMUST00000060232.8
|
Rab2a
|
RAB2A, member RAS oncogene family |
| chr11_+_87017878 | 0.22 |
ENSMUST00000041282.13
|
Trim37
|
tripartite motif-containing 37 |
| chr19_+_8848924 | 0.22 |
ENSMUST00000238036.2
|
Ubxn1
|
UBX domain protein 1 |
| chrX_-_133501677 | 0.22 |
ENSMUST00000239113.2
|
Gla
|
galactosidase, alpha |
| chr12_+_3941728 | 0.22 |
ENSMUST00000172689.8
ENSMUST00000111186.8 |
Dnmt3a
|
DNA methyltransferase 3A |
| chrX_-_73416824 | 0.21 |
ENSMUST00000178691.2
ENSMUST00000114146.8 |
Ubl4a
Slc10a3
|
ubiquitin-like 4A solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr19_+_8848876 | 0.21 |
ENSMUST00000166407.9
|
Ubxn1
|
UBX domain protein 1 |
| chr8_+_86219191 | 0.21 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr8_+_106895488 | 0.21 |
ENSMUST00000034378.5
ENSMUST00000212421.2 |
Slc7a6
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 |
| chr17_+_57586094 | 0.21 |
ENSMUST00000169220.9
ENSMUST00000005889.16 ENSMUST00000112870.5 |
Vav1
|
vav 1 oncogene |
| chr6_+_17694153 | 0.21 |
ENSMUST00000115418.8
|
St7
|
suppression of tumorigenicity 7 |
| chr8_-_34614187 | 0.21 |
ENSMUST00000033910.9
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr8_+_107783502 | 0.20 |
ENSMUST00000034392.13
ENSMUST00000170962.3 |
Nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr3_+_51323383 | 0.20 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr1_-_170002444 | 0.20 |
ENSMUST00000111351.10
ENSMUST00000027981.8 |
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr4_-_135714465 | 0.20 |
ENSMUST00000105851.9
|
Pithd1
|
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr9_-_107167046 | 0.20 |
ENSMUST00000035194.8
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr4_+_130001349 | 0.20 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr16_-_19801781 | 0.20 |
ENSMUST00000058839.10
|
Klhl6
|
kelch-like 6 |
| chr7_-_110682204 | 0.20 |
ENSMUST00000161051.8
ENSMUST00000160132.8 ENSMUST00000162415.9 |
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr11_-_69811347 | 0.20 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_-_103588487 | 0.20 |
ENSMUST00000107013.3
|
Gosr2
|
golgi SNAP receptor complex member 2 |
| chrX_-_93166992 | 0.20 |
ENSMUST00000088102.12
ENSMUST00000113927.8 |
Zfx
|
zinc finger protein X-linked |
| chr3_+_40699763 | 0.19 |
ENSMUST00000203353.3
|
Hspa4l
|
heat shock protein 4 like |
| chr13_-_38178059 | 0.19 |
ENSMUST00000225319.2
ENSMUST00000225246.2 ENSMUST00000021864.8 |
Ssr1
|
signal sequence receptor, alpha |
| chr1_-_170002526 | 0.19 |
ENSMUST00000111350.10
|
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr19_+_6135013 | 0.19 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr15_-_97664931 | 0.19 |
ENSMUST00000177352.8
|
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr8_-_68270870 | 0.19 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_+_129854256 | 0.19 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr1_+_155916655 | 0.19 |
ENSMUST00000065648.15
ENSMUST00000097526.3 |
Tor1aip2
|
torsin A interacting protein 2 |
| chr9_-_36678868 | 0.19 |
ENSMUST00000217599.2
ENSMUST00000120381.9 |
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr1_+_135060994 | 0.19 |
ENSMUST00000167080.3
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr9_+_44238089 | 0.18 |
ENSMUST00000054708.5
|
Dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr15_+_34082805 | 0.18 |
ENSMUST00000022865.17
|
Mtdh
|
metadherin |
| chr11_+_87938519 | 0.18 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr5_-_145138639 | 0.18 |
ENSMUST00000162220.2
ENSMUST00000031632.9 ENSMUST00000198959.2 |
Zkscan14
|
zinc finger with KRAB and SCAN domains 14 |
| chr11_+_87018079 | 0.18 |
ENSMUST00000139532.2
|
Trim37
|
tripartite motif-containing 37 |
| chr10_-_43777712 | 0.18 |
ENSMUST00000020012.7
|
Qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr10_+_79613083 | 0.18 |
ENSMUST00000020575.5
|
Fstl3
|
follistatin-like 3 |
| chr9_-_50256268 | 0.18 |
ENSMUST00000076364.6
|
Rpl10-ps3
|
ribosomal protein L10, pseudogene 3 |
| chr6_+_17693941 | 0.18 |
ENSMUST00000115420.8
ENSMUST00000115419.8 |
St7
|
suppression of tumorigenicity 7 |
| chr11_-_20781009 | 0.17 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr1_-_89858901 | 0.17 |
ENSMUST00000036954.9
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr13_-_36918424 | 0.17 |
ENSMUST00000037623.15
|
Nrn1
|
neuritin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.4 | 2.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 0.8 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.2 | 0.7 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 0.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 0.5 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.2 | 1.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.2 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.7 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.7 | GO:0036166 | phenotypic switching(GO:0036166) cellular response to cocaine(GO:0071314) |
| 0.1 | 0.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.4 | GO:0010269 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.1 | 0.9 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.2 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.8 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 | 0.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 1.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.5 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.4 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.1 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.1 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.4 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.2 | GO:0097212 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.2 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.2 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.5 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 0.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.0 | 0.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.4 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.0 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0021660 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) glycosphingolipid catabolic process(GO:0046479) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.7 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.6 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.4 | GO:0060746 | parental behavior(GO:0060746) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.2 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.3 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.3 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0097144 | BAX complex(GO:0097144) |
| 0.2 | 0.8 | GO:0071920 | cleavage body(GO:0071920) |
| 0.2 | 0.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.2 | 2.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.7 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.0 | 0.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.9 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.2 | 0.5 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.2 | 0.8 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 1.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 2.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.7 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.5 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.8 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.0 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.4 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 1.0 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.0 | 0.2 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.0 | 0.2 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.7 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0031370 | eukaryotic initiation factor 4G binding(GO:0031370) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.8 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.8 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 1.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |