Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Atf7_E4f1
Z-value: 0.53
Transcription factors associated with Atf7_E4f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
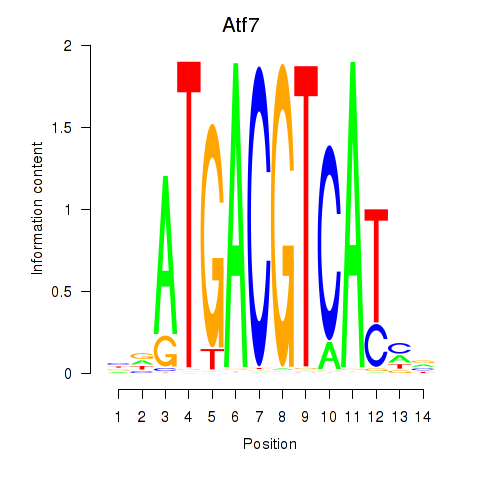
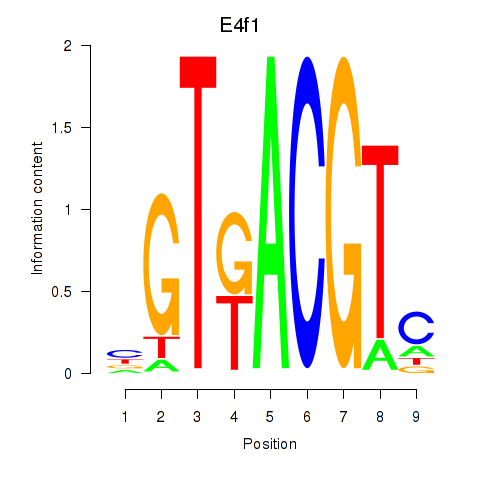
Atf7
|
ENSMUSG00000099083.8 | activating transcription factor 7 |
|
E4f1
|
ENSMUSG00000024137.10 | E4F transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf7 | mm39_v1_chr15_-_102533494_102533535 | 0.62 | 2.6e-01 | Click! |
| E4f1 | mm39_v1_chr17_-_24674252_24674366 | 0.40 | 5.1e-01 | Click! |
Activity profile of Atf7_E4f1 motif
Sorted Z-values of Atf7_E4f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23945189 | 2.41 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr13_+_23715220 | 1.46 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr1_-_71692320 | 1.21 |
ENSMUST00000186940.7
ENSMUST00000188894.7 ENSMUST00000188674.7 ENSMUST00000189821.7 ENSMUST00000187938.7 ENSMUST00000190780.7 ENSMUST00000186736.2 ENSMUST00000055226.13 ENSMUST00000186129.7 |
Fn1
|
fibronectin 1 |
| chr19_-_61216834 | 0.72 |
ENSMUST00000076046.7
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr13_-_21937997 | 0.71 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr13_+_22227359 | 0.66 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr19_-_61215743 | 0.52 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr7_+_44146029 | 0.51 |
ENSMUST00000205359.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr13_+_21938258 | 0.51 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr13_-_54836059 | 0.49 |
ENSMUST00000122935.2
ENSMUST00000128257.8 |
Rnf44
|
ring finger protein 44 |
| chr13_-_67454476 | 0.43 |
ENSMUST00000049705.8
|
Zfp457
|
zinc finger protein 457 |
| chr6_-_39702127 | 0.40 |
ENSMUST00000101497.4
|
Braf
|
Braf transforming gene |
| chr2_+_30127692 | 0.38 |
ENSMUST00000113654.8
ENSMUST00000095078.3 |
Lrrc8a
|
leucine rich repeat containing 8A VRAC subunit A |
| chr17_-_26727437 | 0.34 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr17_+_35200823 | 0.34 |
ENSMUST00000114011.11
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_-_150736554 | 0.34 |
ENSMUST00000117997.2
ENSMUST00000037827.10 |
Slc45a1
|
solute carrier family 45, member 1 |
| chr7_+_16716989 | 0.33 |
ENSMUST00000206129.3
|
Gm42372
|
predicted gene, 42372 |
| chr6_-_149090146 | 0.33 |
ENSMUST00000095319.10
ENSMUST00000141346.2 ENSMUST00000111535.8 |
Amn1
|
antagonist of mitotic exit network 1 |
| chr13_-_54835996 | 0.32 |
ENSMUST00000150806.8
ENSMUST00000125927.8 |
Rnf44
|
ring finger protein 44 |
| chr8_-_26087475 | 0.31 |
ENSMUST00000210810.2
ENSMUST00000210616.2 ENSMUST00000079160.8 |
Letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_+_155118217 | 0.30 |
ENSMUST00000029128.4
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr13_-_54835878 | 0.28 |
ENSMUST00000125871.8
|
Rnf44
|
ring finger protein 44 |
| chr4_+_115594951 | 0.27 |
ENSMUST00000106522.9
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr5_+_123532819 | 0.26 |
ENSMUST00000111596.8
ENSMUST00000068237.12 |
Mlxip
|
MLX interacting protein |
| chr7_+_46700349 | 0.26 |
ENSMUST00000010451.8
|
Tmem86a
|
transmembrane protein 86A |
| chrX_-_20787150 | 0.25 |
ENSMUST00000081893.7
ENSMUST00000115345.8 |
Syn1
|
synapsin I |
| chr17_+_21503204 | 0.24 |
ENSMUST00000236545.2
ENSMUST00000235527.2 |
Vmn1r236
|
vomeronasal 1 receptor 236 |
| chr10_+_121575819 | 0.24 |
ENSMUST00000065600.8
ENSMUST00000136432.2 |
BC048403
|
cDNA sequence BC048403 |
| chr7_-_44145830 | 0.23 |
ENSMUST00000118515.9
ENSMUST00000138328.3 ENSMUST00000239015.2 ENSMUST00000118808.9 |
Emc10
|
ER membrane protein complex subunit 10 |
| chr7_+_112278520 | 0.22 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr8_+_84793453 | 0.22 |
ENSMUST00000211046.2
ENSMUST00000005600.6 |
Rfx1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr13_-_54836077 | 0.21 |
ENSMUST00000150626.2
ENSMUST00000134177.8 |
Rnf44
|
ring finger protein 44 |
| chr19_-_5135510 | 0.20 |
ENSMUST00000140389.8
ENSMUST00000151413.2 ENSMUST00000077066.8 |
Tmem151a
|
transmembrane protein 151A |
| chr2_-_58990967 | 0.20 |
ENSMUST00000226455.2
ENSMUST00000077687.6 |
Ccdc148
|
coiled-coil domain containing 148 |
| chr7_-_12768486 | 0.20 |
ENSMUST00000211344.2
ENSMUST00000210587.2 ENSMUST00000211626.2 ENSMUST00000005711.6 |
Chmp2a
|
charged multivesicular body protein 2A |
| chr4_+_85123654 | 0.19 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr15_-_81284244 | 0.19 |
ENSMUST00000172107.8
ENSMUST00000169204.2 ENSMUST00000163382.2 |
St13
|
suppression of tumorigenicity 13 |
| chr9_+_124196337 | 0.19 |
ENSMUST00000188509.2
|
Ppp2r3d
|
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
| chr8_+_80438421 | 0.19 |
ENSMUST00000048147.9
ENSMUST00000210812.2 |
Anapc10
|
anaphase promoting complex subunit 10 |
| chr17_+_35201130 | 0.18 |
ENSMUST00000173004.2
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr11_+_87295860 | 0.18 |
ENSMUST00000060835.12
|
Tex14
|
testis expressed gene 14 |
| chr19_+_8828132 | 0.17 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr5_-_65650133 | 0.17 |
ENSMUST00000196121.2
|
Gm43552
|
predicted gene 43552 |
| chr11_+_68986043 | 0.17 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr4_+_115594918 | 0.16 |
ENSMUST00000106525.9
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr7_-_26895561 | 0.16 |
ENSMUST00000122202.8
ENSMUST00000080356.10 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr10_-_120037464 | 0.16 |
ENSMUST00000020448.11
|
Irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr19_+_45036220 | 0.16 |
ENSMUST00000084493.8
|
Sfxn3
|
sideroflexin 3 |
| chr4_-_131802606 | 0.16 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr16_-_23807602 | 0.16 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr13_-_67508655 | 0.15 |
ENSMUST00000081582.8
|
Zfp953
|
zinc finger protein 953 |
| chr7_-_101859308 | 0.15 |
ENSMUST00000070165.7
ENSMUST00000211235.2 ENSMUST00000211022.2 |
Nup98
|
nucleoporin 98 |
| chr8_-_112356957 | 0.15 |
ENSMUST00000070004.4
|
Ldhd
|
lactate dehydrogenase D |
| chr17_+_37269468 | 0.15 |
ENSMUST00000040177.7
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr19_+_45036037 | 0.15 |
ENSMUST00000062213.13
|
Sfxn3
|
sideroflexin 3 |
| chr8_-_84996976 | 0.15 |
ENSMUST00000005120.12
ENSMUST00000163993.2 ENSMUST00000098578.10 |
Ccdc130
|
coiled-coil domain containing 130 |
| chr11_+_110858842 | 0.15 |
ENSMUST00000180023.8
ENSMUST00000106636.8 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr4_+_132903646 | 0.15 |
ENSMUST00000105912.2
|
Wasf2
|
WASP family, member 2 |
| chr17_-_33979280 | 0.15 |
ENSMUST00000173860.8
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr7_+_140641010 | 0.15 |
ENSMUST00000048002.7
|
B4galnt4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4 |
| chr17_-_34043320 | 0.14 |
ENSMUST00000173879.8
ENSMUST00000166693.3 ENSMUST00000173019.8 |
Rps28
|
ribosomal protein S28 |
| chr7_+_125307116 | 0.14 |
ENSMUST00000148701.4
|
Katnip
|
katanin interacting protein |
| chr11_-_23720953 | 0.14 |
ENSMUST00000102864.5
|
Rel
|
reticuloendotheliosis oncogene |
| chr2_-_168072295 | 0.14 |
ENSMUST00000154111.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr2_-_164753480 | 0.14 |
ENSMUST00000041361.14
|
Zfp335
|
zinc finger protein 335 |
| chr8_+_4375212 | 0.14 |
ENSMUST00000127460.8
ENSMUST00000136191.8 |
Ccl25
|
chemokine (C-C motif) ligand 25 |
| chr9_+_48406706 | 0.14 |
ENSMUST00000048824.9
|
Gm5617
|
predicted gene 5617 |
| chr18_-_62313019 | 0.14 |
ENSMUST00000053640.5
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr17_-_32074754 | 0.14 |
ENSMUST00000024839.6
|
Sik1
|
salt inducible kinase 1 |
| chr8_+_71207326 | 0.14 |
ENSMUST00000110093.9
ENSMUST00000143118.3 ENSMUST00000034301.12 ENSMUST00000110090.8 |
Rab3a
|
RAB3A, member RAS oncogene family |
| chr11_-_70350783 | 0.13 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr6_+_125026865 | 0.13 |
ENSMUST00000112413.8
|
Acrbp
|
proacrosin binding protein |
| chr11_+_54413772 | 0.13 |
ENSMUST00000207429.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr11_-_70578775 | 0.13 |
ENSMUST00000036299.14
ENSMUST00000119120.2 ENSMUST00000100933.10 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr19_+_59249316 | 0.13 |
ENSMUST00000026084.5
|
Slc18a2
|
solute carrier family 18 (vesicular monoamine), member 2 |
| chr16_+_57369595 | 0.13 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr7_-_73187369 | 0.13 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr10_-_30076488 | 0.13 |
ENSMUST00000216853.2
|
Cenpw
|
centromere protein W |
| chr16_+_17269845 | 0.13 |
ENSMUST00000006293.5
ENSMUST00000231228.2 |
Crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr4_-_89229256 | 0.13 |
ENSMUST00000097981.6
|
Cdkn2b
|
cyclin dependent kinase inhibitor 2B |
| chr7_+_128125339 | 0.13 |
ENSMUST00000033136.9
|
Bag3
|
BCL2-associated athanogene 3 |
| chr2_+_31864438 | 0.13 |
ENSMUST00000065398.13
|
Nup214
|
nucleoporin 214 |
| chr2_-_104324035 | 0.13 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr13_-_38221045 | 0.12 |
ENSMUST00000089840.5
|
Cage1
|
cancer antigen 1 |
| chr7_+_119495058 | 0.12 |
ENSMUST00000106518.9
ENSMUST00000207270.2 ENSMUST00000208424.2 ENSMUST00000208202.2 ENSMUST00000054440.11 |
Lyrm1
|
LYR motif containing 1 |
| chr6_+_82379768 | 0.12 |
ENSMUST00000203775.2
|
Tacr1
|
tachykinin receptor 1 |
| chr7_-_45367382 | 0.12 |
ENSMUST00000107737.11
|
Sphk2
|
sphingosine kinase 2 |
| chr7_-_126992776 | 0.12 |
ENSMUST00000165495.2
ENSMUST00000106303.3 ENSMUST00000074249.7 |
E430018J23Rik
|
RIKEN cDNA E430018J23 gene |
| chr11_-_70578744 | 0.12 |
ENSMUST00000108545.9
ENSMUST00000120261.8 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr4_+_32238712 | 0.11 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr3_+_127347099 | 0.11 |
ENSMUST00000043108.9
ENSMUST00000196141.5 ENSMUST00000195955.2 |
Zgrf1
|
zinc finger, GRF-type containing 1 |
| chr11_+_69214789 | 0.11 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr4_+_40269563 | 0.11 |
ENSMUST00000129758.3
|
Smim27
|
small integral membrane protein 27 |
| chr11_+_54413698 | 0.11 |
ENSMUST00000108895.8
ENSMUST00000101206.10 |
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr10_+_29087602 | 0.11 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr18_-_35348049 | 0.11 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr7_+_139616298 | 0.11 |
ENSMUST00000168194.3
ENSMUST00000210882.2 |
Zfp511
|
zinc finger protein 511 |
| chr17_-_67661382 | 0.11 |
ENSMUST00000223982.2
ENSMUST00000224091.2 |
Ptprm
|
protein tyrosine phosphatase, receptor type, M |
| chr11_-_70578905 | 0.11 |
ENSMUST00000108544.8
|
Camta2
|
calmodulin binding transcription activator 2 |
| chr6_+_124908389 | 0.11 |
ENSMUST00000180095.4
|
Mlf2
|
myeloid leukemia factor 2 |
| chr7_+_121666388 | 0.11 |
ENSMUST00000033158.6
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr6_+_124908439 | 0.11 |
ENSMUST00000032214.14
|
Mlf2
|
myeloid leukemia factor 2 |
| chr18_+_36481706 | 0.11 |
ENSMUST00000235864.2
ENSMUST00000050584.10 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr19_+_45035942 | 0.10 |
ENSMUST00000237222.2
ENSMUST00000111954.11 |
Sfxn3
|
sideroflexin 3 |
| chr7_+_125307060 | 0.10 |
ENSMUST00000124223.8
ENSMUST00000069660.13 |
Katnip
|
katanin interacting protein |
| chr1_+_37338964 | 0.10 |
ENSMUST00000027287.11
ENSMUST00000140264.8 |
Inpp4a
|
inositol polyphosphate-4-phosphatase, type I |
| chr5_+_72360631 | 0.10 |
ENSMUST00000169617.3
|
Atp10d
|
ATPase, class V, type 10D |
| chr16_-_4608084 | 0.10 |
ENSMUST00000118703.8
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr1_+_33947250 | 0.10 |
ENSMUST00000183034.5
|
Dst
|
dystonin |
| chr8_-_23143422 | 0.10 |
ENSMUST00000033938.7
|
Polb
|
polymerase (DNA directed), beta |
| chr7_+_141027557 | 0.10 |
ENSMUST00000106004.8
|
Rplp2
|
ribosomal protein, large P2 |
| chr17_-_43187280 | 0.10 |
ENSMUST00000024709.9
ENSMUST00000233476.2 |
Cd2ap
|
CD2-associated protein |
| chr5_-_139470169 | 0.10 |
ENSMUST00000150992.2
ENSMUST00000110851.8 ENSMUST00000079996.13 |
Zfand2a
|
zinc finger, AN1-type domain 2A |
| chr4_+_150366028 | 0.10 |
ENSMUST00000105682.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr17_-_84773544 | 0.10 |
ENSMUST00000047524.10
|
Thada
|
thyroid adenoma associated |
| chr7_-_45367314 | 0.10 |
ENSMUST00000210060.2
|
Sphk2
|
sphingosine kinase 2 |
| chr9_+_64086553 | 0.10 |
ENSMUST00000034965.8
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr11_+_69214971 | 0.10 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr13_-_100881117 | 0.09 |
ENSMUST00000078573.5
ENSMUST00000109333.8 |
Mrps36
|
mitochondrial ribosomal protein S36 |
| chr5_-_25428151 | 0.09 |
ENSMUST00000066954.2
|
E130116L18Rik
|
RIKEN cDNA E130116L18 gene |
| chr7_-_101859379 | 0.09 |
ENSMUST00000210682.2
|
Nup98
|
nucleoporin 98 |
| chr5_-_86320599 | 0.09 |
ENSMUST00000039373.14
|
Uba6
|
ubiquitin-like modifier activating enzyme 6 |
| chr15_+_12117899 | 0.09 |
ENSMUST00000122941.8
|
Zfr
|
zinc finger RNA binding protein |
| chr10_-_80895949 | 0.09 |
ENSMUST00000219401.2
ENSMUST00000220317.2 ENSMUST00000005067.6 ENSMUST00000218208.2 |
Sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr18_+_82932747 | 0.09 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr7_+_44146012 | 0.09 |
ENSMUST00000205422.2
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr10_+_105676925 | 0.09 |
ENSMUST00000020049.9
|
Ccdc59
|
coiled-coil domain containing 59 |
| chr13_+_38010879 | 0.09 |
ENSMUST00000149745.8
|
Rreb1
|
ras responsive element binding protein 1 |
| chr17_-_56783462 | 0.09 |
ENSMUST00000067538.6
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr9_-_97915036 | 0.09 |
ENSMUST00000162295.2
|
Clstn2
|
calsyntenin 2 |
| chr6_-_34887530 | 0.09 |
ENSMUST00000149448.8
ENSMUST00000133336.8 |
Wdr91
|
WD repeat domain 91 |
| chr16_-_31767250 | 0.09 |
ENSMUST00000202722.2
|
0610012G03Rik
|
RIKEN cDNA 0610012G03 gene |
| chr17_-_56783376 | 0.09 |
ENSMUST00000223859.2
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr15_-_58953838 | 0.09 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr17_-_33979442 | 0.09 |
ENSMUST00000057373.14
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr1_+_59952131 | 0.09 |
ENSMUST00000036540.12
|
Fam117b
|
family with sequence similarity 117, member B |
| chr15_+_79232137 | 0.09 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr1_+_39026887 | 0.09 |
ENSMUST00000194552.2
|
Pdcl3
|
phosducin-like 3 |
| chr8_-_116434517 | 0.09 |
ENSMUST00000109104.2
|
Maf
|
avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr11_+_69805005 | 0.09 |
ENSMUST00000057884.6
|
Gps2
|
G protein pathway suppressor 2 |
| chr4_-_41774097 | 0.08 |
ENSMUST00000108036.8
ENSMUST00000108037.9 ENSMUST00000108032.3 ENSMUST00000173865.9 ENSMUST00000155240.2 |
Ccl27a
|
chemokine (C-C motif) ligand 27A |
| chr2_+_19662529 | 0.08 |
ENSMUST00000052168.6
|
Otud1
|
OTU domain containing 1 |
| chr8_+_3671528 | 0.08 |
ENSMUST00000156380.4
|
Pet100
|
PET100 homolog |
| chr11_+_68582731 | 0.08 |
ENSMUST00000102611.10
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr3_+_106628987 | 0.08 |
ENSMUST00000130105.2
|
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr11_+_51153941 | 0.08 |
ENSMUST00000093132.13
ENSMUST00000109113.8 |
Clk4
|
CDC like kinase 4 |
| chr6_-_30304512 | 0.08 |
ENSMUST00000094543.3
ENSMUST00000102993.10 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr7_+_44145987 | 0.08 |
ENSMUST00000107927.5
|
Fam71e1
|
family with sequence similarity 71, member E1 |
| chr2_+_48839505 | 0.08 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr3_+_45332831 | 0.08 |
ENSMUST00000193252.2
ENSMUST00000171554.8 ENSMUST00000166126.7 ENSMUST00000170695.4 |
Pcdh10
|
protocadherin 10 |
| chr15_-_51728893 | 0.08 |
ENSMUST00000022925.10
|
Eif3h
|
eukaryotic translation initiation factor 3, subunit H |
| chr7_+_4140474 | 0.08 |
ENSMUST00000154571.7
|
Leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chr3_+_106629209 | 0.08 |
ENSMUST00000106736.3
ENSMUST00000154973.2 ENSMUST00000098750.5 ENSMUST00000150513.2 |
Lrif1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr16_+_21613068 | 0.08 |
ENSMUST00000211443.2
ENSMUST00000231300.2 ENSMUST00000209449.2 ENSMUST00000181780.9 ENSMUST00000209728.2 ENSMUST00000181960.3 ENSMUST00000209429.2 ENSMUST00000180830.3 ENSMUST00000231988.2 |
1300002E11Rik
Map3k13
|
RIKEN cDNA 1300002E11 gene mitogen-activated protein kinase kinase kinase 13 |
| chr10_-_61288437 | 0.08 |
ENSMUST00000167087.2
ENSMUST00000020288.15 |
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chrM_+_5319 | 0.08 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr1_-_155912159 | 0.08 |
ENSMUST00000097527.10
|
Tor1aip1
|
torsin A interacting protein 1 |
| chrX_+_150303650 | 0.08 |
ENSMUST00000112666.8
ENSMUST00000112662.9 ENSMUST00000168501.8 |
Phf8
|
PHD finger protein 8 |
| chr11_+_69214883 | 0.08 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr4_-_46138667 | 0.08 |
ENSMUST00000147837.8
|
Tstd2
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
| chr17_+_18372149 | 0.08 |
ENSMUST00000169686.3
|
Vmn2r92
|
vomeronasal 2, receptor 92 |
| chr5_-_125466240 | 0.07 |
ENSMUST00000108707.3
|
Ubc
|
ubiquitin C |
| chr2_+_126549239 | 0.07 |
ENSMUST00000028841.14
ENSMUST00000110416.3 |
Usp8
|
ubiquitin specific peptidase 8 |
| chr15_+_98972850 | 0.07 |
ENSMUST00000039665.8
|
Troap
|
trophinin associated protein |
| chr11_+_54413673 | 0.07 |
ENSMUST00000102743.10
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr13_+_42205491 | 0.07 |
ENSMUST00000060148.6
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr7_+_44398008 | 0.07 |
ENSMUST00000171821.8
|
Vrk3
|
vaccinia related kinase 3 |
| chr7_+_4140031 | 0.07 |
ENSMUST00000128756.8
ENSMUST00000132086.8 ENSMUST00000037472.13 ENSMUST00000117274.8 ENSMUST00000121270.8 ENSMUST00000144248.3 |
Leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chr1_-_134006847 | 0.07 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr7_-_16348862 | 0.07 |
ENSMUST00000171937.2
ENSMUST00000075845.11 |
Arhgap35
|
Rho GTPase activating protein 35 |
| chr16_-_4607848 | 0.07 |
ENSMUST00000004173.12
|
Cdip1
|
cell death inducing Trp53 target 1 |
| chr8_-_3517617 | 0.07 |
ENSMUST00000111081.10
ENSMUST00000004686.13 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr2_-_168072493 | 0.07 |
ENSMUST00000109193.8
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr7_-_126497421 | 0.07 |
ENSMUST00000121532.8
ENSMUST00000032926.12 |
Tmem219
|
transmembrane protein 219 |
| chr11_-_6556053 | 0.07 |
ENSMUST00000045713.4
|
Nacad
|
NAC alpha domain containing |
| chr15_+_79231720 | 0.07 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr18_-_75830595 | 0.07 |
ENSMUST00000165559.3
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr13_+_75855695 | 0.07 |
ENSMUST00000222194.2
ENSMUST00000223535.2 ENSMUST00000222853.2 |
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr11_+_93886906 | 0.07 |
ENSMUST00000041956.14
|
Spag9
|
sperm associated antigen 9 |
| chr9_+_64718708 | 0.06 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr12_+_73333553 | 0.06 |
ENSMUST00000140523.8
ENSMUST00000126488.8 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr17_+_21165573 | 0.06 |
ENSMUST00000007708.14
|
Ppp2r1a
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr11_+_79230618 | 0.06 |
ENSMUST00000219057.2
ENSMUST00000108251.9 ENSMUST00000071325.9 |
Nf1
|
neurofibromin 1 |
| chr17_+_85397980 | 0.06 |
ENSMUST00000095188.7
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr1_-_183126875 | 0.06 |
ENSMUST00000195233.3
|
Mia3
|
melanoma inhibitory activity 3 |
| chr5_-_122752508 | 0.06 |
ENSMUST00000127220.8
ENSMUST00000031426.14 |
Ift81
|
intraflagellar transport 81 |
| chr4_-_131802561 | 0.06 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr5_+_9150691 | 0.06 |
ENSMUST00000115365.3
|
Tmem243
|
transmembrane protein 243, mitochondrial |
| chr19_-_59334172 | 0.06 |
ENSMUST00000099274.4
|
Pdzd8
|
PDZ domain containing 8 |
| chr4_+_43441939 | 0.06 |
ENSMUST00000060864.13
|
Tesk1
|
testis specific protein kinase 1 |
| chr1_+_155911518 | 0.06 |
ENSMUST00000133152.2
|
Tor1aip2
|
torsin A interacting protein 2 |
| chr11_+_69214895 | 0.06 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr11_+_6150029 | 0.06 |
ENSMUST00000181545.2
|
A730071L15Rik
|
RIKEN cDNA A730071L15Rik gene |
| chr6_+_54658609 | 0.06 |
ENSMUST00000190641.7
ENSMUST00000187701.2 |
Mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr1_-_155912216 | 0.06 |
ENSMUST00000027738.14
|
Tor1aip1
|
torsin A interacting protein 1 |
| chr11_-_98329782 | 0.06 |
ENSMUST00000002655.8
|
Mien1
|
migration and invasion enhancer 1 |
| chr17_-_35954573 | 0.06 |
ENSMUST00000095467.4
|
Mucl3
|
mucin like 3 |
| chr17_-_36290571 | 0.06 |
ENSMUST00000173724.2
ENSMUST00000172900.8 ENSMUST00000174849.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr12_+_24701273 | 0.06 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr14_-_87378641 | 0.06 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf7_E4f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 1.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.2 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.0 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.2 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.2 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) amino acid homeostasis(GO:0080144) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.0 | 0.1 | GO:0061188 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.0 | GO:2000328 | regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:1902164 | positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.0 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0033128 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.0 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.0 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 1.2 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.2 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0031711 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.0 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |