Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
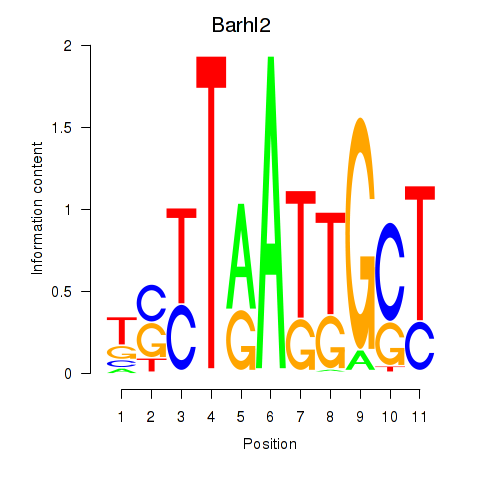
Results for Barhl2
Z-value: 0.97
Transcription factors associated with Barhl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Barhl2
|
ENSMUSG00000034384.12 | BarH like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl2 | mm39_v1_chr5_-_106606032_106606032 | -0.11 | 8.6e-01 | Click! |
Activity profile of Barhl2 motif
Sorted Z-values of Barhl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_24069680 | 0.69 |
ENSMUST00000205428.2
ENSMUST00000171904.3 ENSMUST00000205626.2 |
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr7_+_43086432 | 0.54 |
ENSMUST00000070518.4
|
Nkg7
|
natural killer cell group 7 sequence |
| chrX_+_138464065 | 0.46 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr4_-_120427449 | 0.44 |
ENSMUST00000030381.8
|
Ctps
|
cytidine 5'-triphosphate synthase |
| chrX_+_168662592 | 0.44 |
ENSMUST00000112105.8
ENSMUST00000078947.12 |
Mid1
|
midline 1 |
| chr1_+_165616250 | 0.43 |
ENSMUST00000161971.8
ENSMUST00000187313.7 ENSMUST00000178336.8 ENSMUST00000005907.12 ENSMUST00000027849.11 |
Cd247
|
CD247 antigen |
| chr5_-_110987441 | 0.37 |
ENSMUST00000145318.2
|
Hscb
|
HscB iron-sulfur cluster co-chaperone |
| chr4_+_132495636 | 0.37 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr10_+_39488930 | 0.37 |
ENSMUST00000019987.7
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr7_+_43086554 | 0.35 |
ENSMUST00000206741.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr4_-_9643636 | 0.34 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr8_+_18896267 | 0.33 |
ENSMUST00000149565.8
ENSMUST00000033847.5 |
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr17_+_35481702 | 0.33 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr5_-_115236354 | 0.32 |
ENSMUST00000100848.3
|
Gm10401
|
predicted gene 10401 |
| chr17_-_25946370 | 0.31 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr4_+_65523223 | 0.31 |
ENSMUST00000050850.14
ENSMUST00000107366.2 |
Trim32
|
tripartite motif-containing 32 |
| chr2_-_3513783 | 0.30 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr18_+_34869412 | 0.30 |
ENSMUST00000105038.3
|
Gm3550
|
predicted gene 3550 |
| chr7_+_3632982 | 0.29 |
ENSMUST00000179769.8
ENSMUST00000008517.13 |
Prpf31
|
pre-mRNA processing factor 31 |
| chr12_+_24701273 | 0.29 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr11_+_95733489 | 0.28 |
ENSMUST00000100532.10
ENSMUST00000036088.11 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr10_+_58230203 | 0.27 |
ENSMUST00000105468.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr5_-_123663440 | 0.25 |
ENSMUST00000197682.6
|
Gm49027
|
predicted gene, 49027 |
| chr5_+_88731366 | 0.25 |
ENSMUST00000199312.5
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr11_+_49327451 | 0.25 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr11_-_95733235 | 0.23 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr5_-_65855511 | 0.23 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr12_+_100745314 | 0.23 |
ENSMUST00000069782.11
|
Dglucy
|
D-glutamate cyclase |
| chr7_+_100145192 | 0.22 |
ENSMUST00000133044.3
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr12_-_34578842 | 0.22 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr17_+_35439810 | 0.22 |
ENSMUST00000118793.2
|
Gm16181
|
predicted gene 16181 |
| chr4_-_43000450 | 0.22 |
ENSMUST00000030164.8
|
Vcp
|
valosin containing protein |
| chr5_+_65288418 | 0.22 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr5_+_24679154 | 0.21 |
ENSMUST00000199856.2
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr14_+_30547541 | 0.21 |
ENSMUST00000006701.8
|
Stimate
|
STIM activating enhancer |
| chrX_+_7688528 | 0.20 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr16_-_92196954 | 0.20 |
ENSMUST00000023672.10
|
Rcan1
|
regulator of calcineurin 1 |
| chr12_+_72488625 | 0.20 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr7_+_43093507 | 0.19 |
ENSMUST00000004729.5
ENSMUST00000206286.2 ENSMUST00000206196.2 ENSMUST00000206411.2 |
Etfb
|
electron transferring flavoprotein, beta polypeptide |
| chr11_+_3599183 | 0.19 |
ENSMUST00000096441.5
|
Morc2a
|
microrchidia 2A |
| chr1_-_67077906 | 0.19 |
ENSMUST00000119559.8
ENSMUST00000149996.2 ENSMUST00000027149.12 ENSMUST00000113979.10 |
Lancl1
|
LanC (bacterial lantibiotic synthetase component C)-like 1 |
| chr7_-_98790275 | 0.19 |
ENSMUST00000037968.10
|
Uvrag
|
UV radiation resistance associated gene |
| chr6_-_34294377 | 0.19 |
ENSMUST00000154655.2
ENSMUST00000102980.11 |
Akr1b3
|
aldo-keto reductase family 1, member B3 (aldose reductase) |
| chr5_+_145141333 | 0.18 |
ENSMUST00000085671.10
ENSMUST00000031601.8 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr8_-_35432783 | 0.18 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr1_+_91178288 | 0.18 |
ENSMUST00000171112.8
ENSMUST00000191533.2 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr16_+_10652910 | 0.18 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr1_+_191449946 | 0.18 |
ENSMUST00000133076.7
ENSMUST00000110855.8 |
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr2_+_59442378 | 0.18 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr2_+_118731860 | 0.18 |
ENSMUST00000036578.7
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr11_-_20282684 | 0.17 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr9_-_110453427 | 0.17 |
ENSMUST00000196876.2
ENSMUST00000035069.14 |
Nradd
|
neurotrophin receptor associated death domain |
| chr2_+_180961507 | 0.17 |
ENSMUST00000098971.11
ENSMUST00000054622.15 ENSMUST00000108814.8 ENSMUST00000048608.16 ENSMUST00000108815.8 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr7_-_12063408 | 0.17 |
ENSMUST00000226701.2
|
Vmn1r83
|
vomeronasal 1 receptor 83 |
| chr15_-_100322934 | 0.17 |
ENSMUST00000123461.8
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr19_-_12018681 | 0.16 |
ENSMUST00000214472.2
|
Olfr1423
|
olfactory receptor 1423 |
| chr2_-_25114660 | 0.16 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr5_+_33815466 | 0.16 |
ENSMUST00000074849.13
ENSMUST00000079534.11 ENSMUST00000201633.2 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr4_+_116078830 | 0.15 |
ENSMUST00000030464.14
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr11_-_120463667 | 0.15 |
ENSMUST00000168360.2
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr16_-_22258469 | 0.15 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr4_-_43710231 | 0.15 |
ENSMUST00000217544.2
ENSMUST00000107862.3 |
Olfr71
|
olfactory receptor 71 |
| chr8_+_94941192 | 0.15 |
ENSMUST00000079961.14
ENSMUST00000212824.2 |
Nup93
|
nucleoporin 93 |
| chr19_-_6947112 | 0.14 |
ENSMUST00000025912.10
|
Plcb3
|
phospholipase C, beta 3 |
| chr5_+_88731386 | 0.14 |
ENSMUST00000031229.11
|
Rufy3
|
RUN and FYVE domain containing 3 |
| chr10_+_58230183 | 0.14 |
ENSMUST00000020077.11
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_-_34356567 | 0.14 |
ENSMUST00000179647.2
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr16_+_36648728 | 0.13 |
ENSMUST00000114819.8
ENSMUST00000023535.4 |
Iqcb1
|
IQ calmodulin-binding motif containing 1 |
| chr7_+_127441119 | 0.13 |
ENSMUST00000121705.8
|
Stx4a
|
syntaxin 4A (placental) |
| chr10_+_18345706 | 0.13 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr6_+_125529911 | 0.12 |
ENSMUST00000112254.8
ENSMUST00000112253.6 |
Vwf
|
Von Willebrand factor |
| chr16_-_3690243 | 0.12 |
ENSMUST00000090522.5
|
Zfp597
|
zinc finger protein 597 |
| chr11_-_40646090 | 0.12 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr1_+_165616315 | 0.12 |
ENSMUST00000161559.3
|
Cd247
|
CD247 antigen |
| chr3_+_96537484 | 0.12 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr1_+_91178017 | 0.11 |
ENSMUST00000059743.12
ENSMUST00000178627.8 ENSMUST00000171165.8 ENSMUST00000191368.7 |
Ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr3_-_54621774 | 0.11 |
ENSMUST00000178832.2
|
Gm21958
|
predicted gene, 21958 |
| chr6_+_57234937 | 0.11 |
ENSMUST00000228297.2
|
Vmn1r15
|
vomeronasal 1 receptor 15 |
| chr13_+_96524802 | 0.11 |
ENSMUST00000099295.6
|
Poc5
|
POC5 centriolar protein |
| chr19_+_24853039 | 0.10 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr6_-_136899167 | 0.10 |
ENSMUST00000032343.7
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr7_-_90125178 | 0.10 |
ENSMUST00000032843.9
|
Tmem126b
|
transmembrane protein 126B |
| chr15_+_76579960 | 0.10 |
ENSMUST00000229679.2
|
Gpt
|
glutamic pyruvic transaminase, soluble |
| chr15_-_100322089 | 0.09 |
ENSMUST00000154331.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr2_+_30282414 | 0.09 |
ENSMUST00000123202.8
ENSMUST00000113612.10 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr2_+_22959223 | 0.09 |
ENSMUST00000114523.10
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5 |
| chr8_-_84831391 | 0.09 |
ENSMUST00000041367.9
ENSMUST00000210279.2 |
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr10_+_127126643 | 0.09 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr11_+_49737558 | 0.08 |
ENSMUST00000109179.9
ENSMUST00000178543.8 ENSMUST00000164643.8 ENSMUST00000020634.14 |
Mapk9
|
mitogen-activated protein kinase 9 |
| chr3_-_69034425 | 0.08 |
ENSMUST00000194558.6
ENSMUST00000029353.9 |
Kpna4
|
karyopherin (importin) alpha 4 |
| chr13_+_51799268 | 0.08 |
ENSMUST00000075853.6
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr18_-_84104574 | 0.08 |
ENSMUST00000175783.3
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr11_-_120464062 | 0.07 |
ENSMUST00000026122.11
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr2_-_120370333 | 0.07 |
ENSMUST00000171215.8
|
Zfp106
|
zinc finger protein 106 |
| chr3_-_129518723 | 0.07 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr7_-_126975169 | 0.07 |
ENSMUST00000205832.2
|
Zfp747
|
zinc finger protein 747 |
| chr11_+_62711295 | 0.07 |
ENSMUST00000108703.2
|
Trim16
|
tripartite motif-containing 16 |
| chr9_-_13738304 | 0.07 |
ENSMUST00000148086.8
ENSMUST00000034398.12 |
Cep57
|
centrosomal protein 57 |
| chr18_+_4993795 | 0.07 |
ENSMUST00000153016.8
|
Svil
|
supervillin |
| chr2_+_180961599 | 0.07 |
ENSMUST00000153112.2
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr4_-_58553553 | 0.07 |
ENSMUST00000107575.9
ENSMUST00000107574.8 ENSMUST00000147354.8 |
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr1_+_171265103 | 0.07 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr18_-_84104507 | 0.06 |
ENSMUST00000060303.10
|
Tshz1
|
teashirt zinc finger family member 1 |
| chr1_+_105918383 | 0.06 |
ENSMUST00000119166.8
|
Zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr14_-_99337137 | 0.06 |
ENSMUST00000042471.11
|
Dis3
|
DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease |
| chr7_+_126810780 | 0.06 |
ENSMUST00000032910.13
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr11_+_70910685 | 0.05 |
ENSMUST00000155236.2
ENSMUST00000143762.2 ENSMUST00000136137.2 |
Mis12
|
MIS12 kinetochore complex component |
| chr16_-_22258320 | 0.05 |
ENSMUST00000170393.2
|
Etv5
|
ets variant 5 |
| chr11_+_62711057 | 0.05 |
ENSMUST00000055006.12
ENSMUST00000072639.10 |
Trim16
|
tripartite motif-containing 16 |
| chr7_+_127440924 | 0.05 |
ENSMUST00000033075.14
|
Stx4a
|
syntaxin 4A (placental) |
| chr7_-_118304930 | 0.05 |
ENSMUST00000207323.2
ENSMUST00000038791.15 |
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr9_-_13738204 | 0.05 |
ENSMUST00000147115.8
|
Cep57
|
centrosomal protein 57 |
| chr7_-_19449319 | 0.05 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr5_+_57875309 | 0.05 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr19_-_4861536 | 0.05 |
ENSMUST00000179909.8
ENSMUST00000172000.3 ENSMUST00000180008.2 ENSMUST00000113793.4 ENSMUST00000006625.8 |
Gm21992
Rbm14
|
predicted gene 21992 RNA binding motif protein 14 |
| chr6_+_8259379 | 0.05 |
ENSMUST00000162034.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr1_+_105918114 | 0.04 |
ENSMUST00000118196.8
|
Zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr10_-_99595498 | 0.04 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr9_-_105008972 | 0.04 |
ENSMUST00000186925.2
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr12_-_108145498 | 0.04 |
ENSMUST00000071095.14
|
Setd3
|
SET domain containing 3 |
| chr7_+_127846121 | 0.04 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr2_+_57887896 | 0.04 |
ENSMUST00000112616.8
ENSMUST00000166729.2 |
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr7_+_12034357 | 0.04 |
ENSMUST00000072801.5
ENSMUST00000227672.2 |
Vmn1r82
|
vomeronasal 1 receptor 82 |
| chr14_+_55797934 | 0.04 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr11_+_95733109 | 0.04 |
ENSMUST00000107714.9
ENSMUST00000107711.8 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr3_-_49711706 | 0.04 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr15_+_101371353 | 0.04 |
ENSMUST00000088049.5
|
Krt86
|
keratin 86 |
| chr12_-_91815855 | 0.04 |
ENSMUST00000167466.2
ENSMUST00000021347.12 ENSMUST00000178462.8 |
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr6_+_8259405 | 0.04 |
ENSMUST00000160705.8
ENSMUST00000159433.8 |
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr3_+_95496270 | 0.03 |
ENSMUST00000176674.8
ENSMUST00000177389.8 ENSMUST00000176755.8 ENSMUST00000177399.2 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr15_-_100321973 | 0.03 |
ENSMUST00000154676.2
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr9_+_39657545 | 0.03 |
ENSMUST00000213358.2
|
Olfr967
|
olfactory receptor 967 |
| chr3_-_49711765 | 0.03 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr7_-_3632826 | 0.03 |
ENSMUST00000205596.2
ENSMUST00000155592.8 ENSMUST00000108641.10 |
Tfpt
|
TCF3 (E2A) fusion partner |
| chr11_-_100504159 | 0.03 |
ENSMUST00000146840.3
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr3_+_95496239 | 0.02 |
ENSMUST00000177390.8
ENSMUST00000060323.12 ENSMUST00000098861.11 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr10_-_107321938 | 0.02 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr7_+_28580847 | 0.02 |
ENSMUST00000066880.6
|
Capn12
|
calpain 12 |
| chr8_+_10299288 | 0.02 |
ENSMUST00000214643.2
|
Myo16
|
myosin XVI |
| chr8_+_57964956 | 0.02 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr6_-_136758716 | 0.01 |
ENSMUST00000078095.11
ENSMUST00000032338.10 |
Gucy2c
|
guanylate cyclase 2c |
| chr8_+_95807814 | 0.01 |
ENSMUST00000034239.9
|
Katnb1
|
katanin p80 (WD40-containing) subunit B 1 |
| chr16_-_45544960 | 0.01 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr11_-_99228756 | 0.01 |
ENSMUST00000100482.3
|
Krt26
|
keratin 26 |
| chr13_-_21765822 | 0.01 |
ENSMUST00000206526.3
ENSMUST00000213912.2 |
Olfr1364
|
olfactory receptor 1364 |
| chr11_+_49231361 | 0.01 |
ENSMUST00000062719.2
|
Olfr1390
|
olfactory receptor 1390 |
| chr14_-_68771138 | 0.01 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr14_+_65504067 | 0.01 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr6_-_8259098 | 0.01 |
ENSMUST00000012627.5
|
Rpa3
|
replication protein A3 |
| chr15_-_101482320 | 0.01 |
ENSMUST00000042957.6
|
Krt75
|
keratin 75 |
| chr4_-_96673423 | 0.01 |
ENSMUST00000107071.2
|
Gm12695
|
predicted gene 12695 |
| chr2_-_57942844 | 0.01 |
ENSMUST00000090940.6
|
Ermn
|
ermin, ERM-like protein |
| chr15_+_66542598 | 0.01 |
ENSMUST00000065916.14
|
Tg
|
thyroglobulin |
| chr6_-_89847511 | 0.01 |
ENSMUST00000089418.5
|
Vmn1r43
|
vomeronasal 1 receptor 43 |
| chr1_-_163552693 | 0.01 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr9_-_105009023 | 0.01 |
ENSMUST00000035179.9
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr8_-_62576140 | 0.01 |
ENSMUST00000034052.14
ENSMUST00000034054.9 |
Anxa10
|
annexin A10 |
| chr6_+_125122172 | 0.01 |
ENSMUST00000119527.8
ENSMUST00000117675.8 |
Iffo1
|
intermediate filament family orphan 1 |
| chr5_+_88523967 | 0.01 |
ENSMUST00000073363.2
|
Amtn
|
amelotin |
| chr5_+_20112704 | 0.00 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr16_-_36648586 | 0.00 |
ENSMUST00000023537.6
ENSMUST00000114829.9 |
Eaf2
|
ELL associated factor 2 |
| chr4_+_116078787 | 0.00 |
ENSMUST00000147292.8
|
Pik3r3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr2_-_85230875 | 0.00 |
ENSMUST00000099927.2
|
Olfr992
|
olfactory receptor 992 |
| chr2_-_87955337 | 0.00 |
ENSMUST00000099833.6
|
Olfr1166
|
olfactory receptor 1166 |
| chr7_-_106473450 | 0.00 |
ENSMUST00000060879.6
|
Olfr705
|
olfactory receptor 705 |
| chr4_-_127224591 | 0.00 |
ENSMUST00000046532.4
|
Gjb3
|
gap junction protein, beta 3 |
| chr11_-_58443063 | 0.00 |
ENSMUST00000073933.2
|
Olfr328
|
olfactory receptor 328 |
| chr2_+_125876883 | 0.00 |
ENSMUST00000110442.2
|
Fgf7
|
fibroblast growth factor 7 |
| chr11_-_99494134 | 0.00 |
ENSMUST00000072306.4
|
Gm11938
|
predicted gene 11938 |
| chr7_-_104542833 | 0.00 |
ENSMUST00000081116.2
|
Olfr666
|
olfactory receptor 666 |
| chr14_+_65504151 | 0.00 |
ENSMUST00000169656.3
ENSMUST00000226005.2 |
Fbxo16
|
F-box protein 16 |
| chr14_+_53007210 | 0.00 |
ENSMUST00000178768.4
|
Trav7d-4
|
T cell receptor alpha variable 7D-4 |
| chr3_-_154034271 | 0.00 |
ENSMUST00000204403.2
|
Lhx8
|
LIM homeobox protein 8 |
| chr2_+_125876566 | 0.00 |
ENSMUST00000064794.14
|
Fgf7
|
fibroblast growth factor 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Barhl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.1 | 0.3 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.1 | 0.2 | GO:0018931 | hexitol metabolic process(GO:0006059) naphthalene metabolic process(GO:0018931) naphthalene-containing compound metabolic process(GO:0090420) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.2 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.3 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.4 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.5 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.3 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.0 | 0.2 | GO:0015193 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |