Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
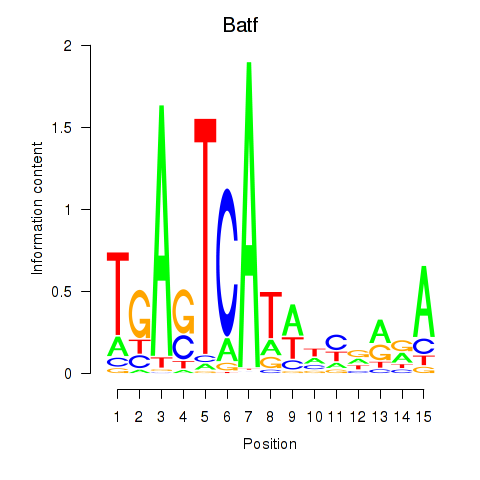
Results for Batf
Z-value: 0.86
Transcription factors associated with Batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf
|
ENSMUSG00000034266.7 | basic leucine zipper transcription factor, ATF-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf | mm39_v1_chr12_+_85733415_85733478 | -0.09 | 8.8e-01 | Click! |
Activity profile of Batf motif
Sorted Z-values of Batf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_21964747 | 1.18 |
ENSMUST00000080511.3
|
H1f5
|
H1.5 linker histone, cluster member |
| chr4_-_118795809 | 0.71 |
ENSMUST00000215312.3
|
Olfr1328
|
olfactory receptor 1328 |
| chrX_-_20955370 | 0.63 |
ENSMUST00000040667.13
|
Zfp300
|
zinc finger protein 300 |
| chr2_+_88470886 | 0.63 |
ENSMUST00000217379.2
ENSMUST00000120598.3 |
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1 |
| chr14_-_32907446 | 0.62 |
ENSMUST00000159606.2
|
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr9_+_75682637 | 0.55 |
ENSMUST00000012281.8
|
Bmp5
|
bone morphogenetic protein 5 |
| chr2_+_88285211 | 0.54 |
ENSMUST00000219871.2
ENSMUST00000213115.3 |
Olfr1183
|
olfactory receptor 1183 |
| chr7_-_85895409 | 0.52 |
ENSMUST00000165771.2
ENSMUST00000233075.2 ENSMUST00000233317.2 ENSMUST00000233312.2 ENSMUST00000233928.2 ENSMUST00000232799.2 ENSMUST00000233744.2 |
Vmn2r76
|
vomeronasal 2, receptor 76 |
| chr18_+_82929451 | 0.50 |
ENSMUST00000235902.2
|
Zfp516
|
zinc finger protein 516 |
| chr9_+_37995368 | 0.50 |
ENSMUST00000212502.4
|
Olfr887
|
olfactory receptor 887 |
| chr11_-_78950698 | 0.47 |
ENSMUST00000141409.8
|
Ksr1
|
kinase suppressor of ras 1 |
| chr1_+_107456731 | 0.46 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr9_+_38026494 | 0.42 |
ENSMUST00000217286.2
|
Olfr889
|
olfactory receptor 889 |
| chr18_+_82928782 | 0.41 |
ENSMUST00000235793.2
|
Zfp516
|
zinc finger protein 516 |
| chr2_+_71884943 | 0.40 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr6_-_122833109 | 0.34 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1 |
| chr2_-_111843053 | 0.33 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr18_+_82928959 | 0.32 |
ENSMUST00000171238.8
|
Zfp516
|
zinc finger protein 516 |
| chr9_+_106247943 | 0.32 |
ENSMUST00000173748.2
|
Dusp7
|
dual specificity phosphatase 7 |
| chr14_+_45588632 | 0.31 |
ENSMUST00000228818.2
|
Styx
|
serine/threonine/tyrosine interaction protein |
| chr16_-_59421342 | 0.28 |
ENSMUST00000172910.3
|
Crybg3
|
beta-gamma crystallin domain containing 3 |
| chr9_+_123921573 | 0.28 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr6_-_97436223 | 0.27 |
ENSMUST00000113359.8
|
Frmd4b
|
FERM domain containing 4B |
| chr11_-_97913420 | 0.24 |
ENSMUST00000103144.10
ENSMUST00000017552.13 ENSMUST00000092736.11 ENSMUST00000107562.2 |
Cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr13_+_120085355 | 0.24 |
ENSMUST00000099241.4
|
Ccl28
|
chemokine (C-C motif) ligand 28 |
| chr12_+_71062733 | 0.23 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr19_-_56810593 | 0.23 |
ENSMUST00000118592.8
|
Ccdc186
|
coiled-coil domain containing 186 |
| chr8_+_109429732 | 0.22 |
ENSMUST00000188994.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chrX_-_104919201 | 0.19 |
ENSMUST00000198209.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr1_-_171476559 | 0.19 |
ENSMUST00000111276.9
ENSMUST00000194531.6 |
Slamf7
|
SLAM family member 7 |
| chr3_-_37286714 | 0.19 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr11_+_75542902 | 0.17 |
ENSMUST00000102504.10
|
Myo1c
|
myosin IC |
| chr18_+_42408418 | 0.17 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr3_-_79439181 | 0.16 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2 |
| chr7_-_12829100 | 0.15 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr12_+_83678987 | 0.15 |
ENSMUST00000048155.16
ENSMUST00000182618.8 ENSMUST00000183154.8 ENSMUST00000182036.8 ENSMUST00000182347.8 |
Rbm25
|
RNA binding motif protein 25 |
| chr13_+_20295672 | 0.14 |
ENSMUST00000180626.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr10_+_128542120 | 0.14 |
ENSMUST00000054125.9
|
Pmel
|
premelanosome protein |
| chr5_-_86666408 | 0.13 |
ENSMUST00000140095.2
ENSMUST00000134179.8 |
Tmprss11g
|
transmembrane protease, serine 11g |
| chr17_+_48080113 | 0.13 |
ENSMUST00000160373.8
ENSMUST00000159641.8 |
Tfeb
|
transcription factor EB |
| chr4_-_155953849 | 0.11 |
ENSMUST00000030950.8
|
Cptp
|
ceramide-1-phosphate transfer protein |
| chrX_-_104918911 | 0.10 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr18_-_38131766 | 0.10 |
ENSMUST00000236588.2
ENSMUST00000237272.2 ENSMUST00000236134.2 |
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr6_-_141892517 | 0.10 |
ENSMUST00000168119.8
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr4_-_123466853 | 0.10 |
ENSMUST00000238555.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr19_-_12093187 | 0.09 |
ENSMUST00000208391.3
ENSMUST00000214103.2 |
Olfr1428
|
olfactory receptor 1428 |
| chr10_+_129219952 | 0.09 |
ENSMUST00000214064.2
|
Olfr784
|
olfactory receptor 784 |
| chr1_+_180978491 | 0.08 |
ENSMUST00000134115.8
ENSMUST00000111059.2 |
Cnih4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr9_-_44179479 | 0.08 |
ENSMUST00000213803.2
|
Nlrx1
|
NLR family member X1 |
| chr17_+_38082190 | 0.08 |
ENSMUST00000217119.2
|
Olfr122
|
olfactory receptor 122 |
| chr11_-_78950641 | 0.06 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr16_-_19241884 | 0.04 |
ENSMUST00000206110.4
|
Olfr165
|
olfactory receptor 165 |
| chr3_+_103034843 | 0.04 |
ENSMUST00000172288.3
|
Dennd2c
|
DENN/MADD domain containing 2C |
| chr5_+_67473069 | 0.04 |
ENSMUST00000202770.2
|
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr14_+_26789345 | 0.03 |
ENSMUST00000226105.2
|
Il17rd
|
interleukin 17 receptor D |
| chr12_-_115258124 | 0.03 |
ENSMUST00000192591.2
|
Ighv8-8
|
immunoglobulin heavy variable 8-8 |
| chr7_-_142215027 | 0.03 |
ENSMUST00000105936.8
|
Igf2
|
insulin-like growth factor 2 |
| chr9_-_39914761 | 0.02 |
ENSMUST00000215523.2
|
Olfr979
|
olfactory receptor 979 |
| chr10_-_117074501 | 0.02 |
ENSMUST00000159193.8
ENSMUST00000020392.5 |
9530003J23Rik
|
RIKEN cDNA 9530003J23 gene |
| chr7_+_102834996 | 0.02 |
ENSMUST00000218618.2
|
Olfr592
|
olfactory receptor 592 |
| chr16_+_35590745 | 0.01 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr2_-_126625343 | 0.01 |
ENSMUST00000028842.9
ENSMUST00000130356.3 |
Usp50
|
ubiquitin specific peptidase 50 |
| chr5_+_137759934 | 0.01 |
ENSMUST00000110983.3
ENSMUST00000031738.5 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr2_-_36995660 | 0.01 |
ENSMUST00000100144.2
|
Olfr362
|
olfactory receptor 362 |
| chrX_+_73636285 | 0.01 |
ENSMUST00000216108.2
ENSMUST00000216007.2 |
Olfr1325
|
olfactory receptor 1325 |
| chr2_-_74409723 | 0.01 |
ENSMUST00000130586.8
|
Lnpk
|
lunapark, ER junction formation factor |
| chr9_+_7272514 | 0.00 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr18_+_82929037 | 0.00 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr7_+_98092628 | 0.00 |
ENSMUST00000098274.5
|
Gucy2d
|
guanylate cyclase 2d |
| chr7_-_102408573 | 0.00 |
ENSMUST00000210453.3
ENSMUST00000232246.3 ENSMUST00000239110.2 ENSMUST00000060187.15 ENSMUST00000168007.3 |
Olfr560
Olfr78
|
olfactory receptor 560 olfactory receptor 78 |
| chr14_+_55459462 | 0.00 |
ENSMUST00000022820.12
|
Dhrs2
|
dehydrogenase/reductase member 2 |
| chr17_-_31383976 | 0.00 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chr12_+_86516875 | 0.00 |
ENSMUST00000167891.2
|
Esrrb
|
estrogen related receptor, beta |
| chrY_-_13465579 | 0.00 |
ENSMUST00000178428.2
|
Gm21866
|
predicted gene, 21866 |
| chr6_-_40917431 | 0.00 |
ENSMUST00000122181.8
ENSMUST00000031935.10 |
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.1 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.4 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.3 | GO:0097394 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 0.3 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.2 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 1.0 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.3 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |