Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
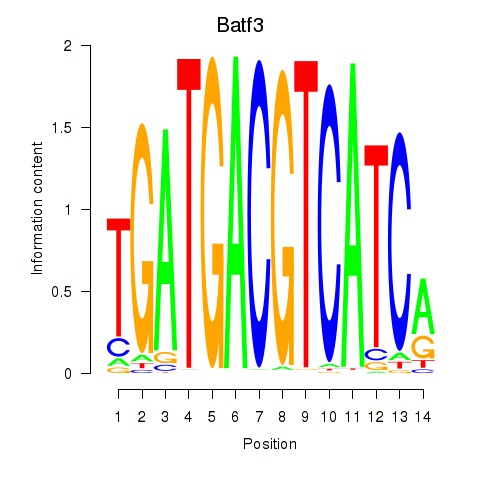
Results for Batf3
Z-value: 0.41
Transcription factors associated with Batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf3
|
ENSMUSG00000026630.10 | basic leucine zipper transcription factor, ATF-like 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Batf3 | mm39_v1_chr1_+_190830522_190830633 | -0.36 | 5.5e-01 | Click! |
Activity profile of Batf3 motif
Sorted Z-values of Batf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_51045182 | 1.60 |
ENSMUST00000227614.2
|
Ccnb1ip1
|
cyclin B1 interacting protein 1 |
| chr13_+_22227359 | 0.89 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr13_-_23945189 | 0.82 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr17_-_26727437 | 0.45 |
ENSMUST00000236661.2
ENSMUST00000025025.7 |
Dusp1
|
dual specificity phosphatase 1 |
| chr13_-_67454476 | 0.42 |
ENSMUST00000049705.8
|
Zfp457
|
zinc finger protein 457 |
| chr13_-_22227114 | 0.41 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr19_-_61215743 | 0.38 |
ENSMUST00000237386.2
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr11_+_69214971 | 0.16 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr11_+_69214789 | 0.15 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr7_+_16716989 | 0.15 |
ENSMUST00000206129.3
|
Gm42372
|
predicted gene, 42372 |
| chr13_-_67508655 | 0.14 |
ENSMUST00000081582.8
|
Zfp953
|
zinc finger protein 953 |
| chr19_+_8828132 | 0.14 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr11_+_69214895 | 0.13 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chr4_+_32238712 | 0.13 |
ENSMUST00000108180.9
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr11_+_69214883 | 0.13 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr4_-_89229256 | 0.12 |
ENSMUST00000097981.6
|
Cdkn2b
|
cyclin dependent kinase inhibitor 2B |
| chr18_-_67682312 | 0.12 |
ENSMUST00000224799.2
|
Spire1
|
spire type actin nucleation factor 1 |
| chr7_+_112278520 | 0.12 |
ENSMUST00000084705.13
ENSMUST00000239442.2 ENSMUST00000239404.2 ENSMUST00000059768.18 |
Tead1
|
TEA domain family member 1 |
| chr14_-_55950939 | 0.10 |
ENSMUST00000168729.8
ENSMUST00000228123.2 ENSMUST00000178034.9 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr16_+_38383182 | 0.10 |
ENSMUST00000163948.8
|
Tmem39a
|
transmembrane protein 39a |
| chr17_-_56783376 | 0.09 |
ENSMUST00000223859.2
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chr4_-_41275091 | 0.08 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr17_-_33979280 | 0.08 |
ENSMUST00000173860.8
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr17_-_33979442 | 0.08 |
ENSMUST00000057373.14
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr15_+_79232137 | 0.08 |
ENSMUST00000163691.3
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr19_-_5135510 | 0.07 |
ENSMUST00000140389.8
ENSMUST00000151413.2 ENSMUST00000077066.8 |
Tmem151a
|
transmembrane protein 151A |
| chr14_-_87378641 | 0.07 |
ENSMUST00000168889.3
ENSMUST00000022599.14 |
Diaph3
|
diaphanous related formin 3 |
| chrX_-_8042129 | 0.07 |
ENSMUST00000143984.2
|
Tbc1d25
|
TBC1 domain family, member 25 |
| chr15_+_79231720 | 0.07 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr5_-_25428151 | 0.06 |
ENSMUST00000066954.2
|
E130116L18Rik
|
RIKEN cDNA E130116L18 gene |
| chr17_-_56185930 | 0.06 |
ENSMUST00000079642.8
|
Zfp119a
|
zinc finger protein 119a |
| chr6_-_86646118 | 0.05 |
ENSMUST00000001184.10
|
Mxd1
|
MAX dimerization protein 1 |
| chr17_-_56252259 | 0.05 |
ENSMUST00000056147.14
|
Zfp119b
|
zinc finger protein 119b |
| chr5_+_25427860 | 0.05 |
ENSMUST00000045737.14
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr6_+_83031502 | 0.04 |
ENSMUST00000092618.9
|
Aup1
|
ancient ubiquitous protein 1 |
| chr12_+_73333553 | 0.04 |
ENSMUST00000140523.8
ENSMUST00000126488.8 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr17_-_36290571 | 0.04 |
ENSMUST00000173724.2
ENSMUST00000172900.8 ENSMUST00000174849.8 |
Prr3
|
proline-rich polypeptide 3 |
| chr11_+_79230618 | 0.04 |
ENSMUST00000219057.2
ENSMUST00000108251.9 ENSMUST00000071325.9 |
Nf1
|
neurofibromin 1 |
| chr4_+_32238950 | 0.03 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr8_+_94537460 | 0.03 |
ENSMUST00000034198.15
ENSMUST00000125716.8 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr16_+_38383154 | 0.03 |
ENSMUST00000171687.8
ENSMUST00000002924.15 |
Tmem39a
|
transmembrane protein 39a |
| chr2_+_162859274 | 0.03 |
ENSMUST00000018002.13
ENSMUST00000150396.3 |
Ift52
|
intraflagellar transport 52 |
| chr5_+_25721059 | 0.03 |
ENSMUST00000045016.9
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr12_-_69205882 | 0.02 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr5_+_130284431 | 0.01 |
ENSMUST00000044204.12
|
Tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr7_-_138447933 | 0.01 |
ENSMUST00000118810.2
ENSMUST00000075667.5 ENSMUST00000119664.2 |
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr3_-_88455556 | 0.01 |
ENSMUST00000131775.2
ENSMUST00000008745.13 |
Rab25
|
RAB25, member RAS oncogene family |
| chr7_-_33065747 | 0.01 |
ENSMUST00000179248.2
|
Scgb2b20
|
secretoglobin, family 2B, member 20 |
| chr9_+_102595628 | 0.01 |
ENSMUST00000156485.2
ENSMUST00000145937.2 ENSMUST00000134483.2 ENSMUST00000190047.7 |
Amotl2
|
angiomotin-like 2 |
| chr7_+_28455563 | 0.01 |
ENSMUST00000178767.3
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr18_+_76944006 | 0.01 |
ENSMUST00000166956.2
|
Skor2
|
SKI family transcriptional corepressor 2 |
| chr7_+_144854542 | 0.00 |
ENSMUST00000033386.12
|
Mrgprf
|
MAS-related GPR, member F |
| chr2_-_35257741 | 0.00 |
ENSMUST00000028243.2
|
4930568D16Rik
|
RIKEN cDNA 4930568D16 gene |
| chr7_-_32873289 | 0.00 |
ENSMUST00000186529.2
|
Scgb2b18
|
secretoglobin, family 2B, member 18 |
| chr7_-_31910570 | 0.00 |
ENSMUST00000188293.2
|
Scgb2b11
|
secretoglobin, family 2B, member 11 |
| chr7_+_144854627 | 0.00 |
ENSMUST00000117718.2
|
Mrgprf
|
MAS-related GPR, member F |
| chr7_+_112278534 | 0.00 |
ENSMUST00000106638.10
|
Tead1
|
TEA domain family member 1 |
| chr16_+_20514925 | 0.00 |
ENSMUST00000128273.2
|
Fam131a
|
family with sequence similarity 131, member A |
| chr19_+_5385672 | 0.00 |
ENSMUST00000043380.5
|
Catsper1
|
cation channel, sperm associated 1 |
| chr15_-_76906150 | 0.00 |
ENSMUST00000230031.2
|
Mb
|
myoglobin |
| chr3_-_19749489 | 0.00 |
ENSMUST00000061294.5
|
Crh
|
corticotropin releasing hormone |
| chr1_-_126758520 | 0.00 |
ENSMUST00000162646.8
|
Nckap5
|
NCK-associated protein 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Batf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.4 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |