Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Brca1
Z-value: 2.22
Transcription factors associated with Brca1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Brca1
|
ENSMUSG00000017146.13 | breast cancer 1, early onset |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Brca1 | mm39_v1_chr11_-_101442663_101442719 | 0.80 | 1.0e-01 | Click! |
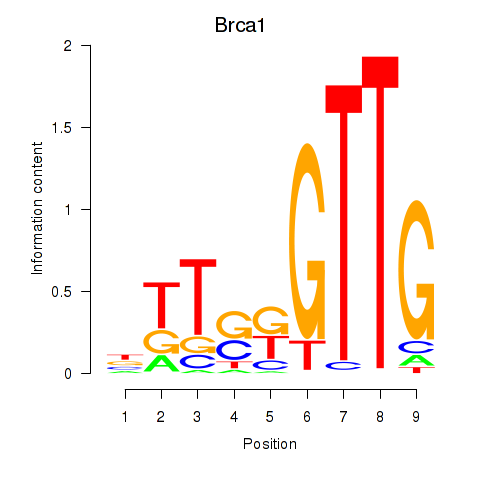
Activity profile of Brca1 motif
Sorted Z-values of Brca1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87684548 | 2.41 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr18_-_43610829 | 2.10 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr13_+_28441511 | 1.60 |
ENSMUST00000223428.2
|
Rps18-ps5
|
ribosomal protein S18, pseudogene 5 |
| chr9_+_88209250 | 1.44 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr7_+_100143250 | 1.39 |
ENSMUST00000153287.8
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr13_-_97897139 | 1.27 |
ENSMUST00000074072.5
|
Rps18-ps6
|
ribosomal protein S18, pseudogene 6 |
| chrX_+_168468186 | 1.15 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr5_+_136982100 | 1.01 |
ENSMUST00000111094.8
ENSMUST00000111097.8 |
Fis1
|
fission, mitochondrial 1 |
| chr8_+_85696453 | 0.96 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr12_+_52144511 | 0.95 |
ENSMUST00000040090.16
|
Nubpl
|
nucleotide binding protein-like |
| chr17_+_6697511 | 0.83 |
ENSMUST00000179569.3
|
Dynlt1b
|
dynein light chain Tctex-type 1B |
| chr11_-_93776580 | 0.82 |
ENSMUST00000066888.10
|
Utp18
|
UTP18 small subunit processome component |
| chr8_+_85696396 | 0.80 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr12_+_111005768 | 0.77 |
ENSMUST00000084968.14
|
Rcor1
|
REST corepressor 1 |
| chr8_+_47070326 | 0.76 |
ENSMUST00000211115.2
ENSMUST00000093517.7 |
Casp3
|
caspase 3 |
| chr5_+_137756407 | 0.73 |
ENSMUST00000141733.8
ENSMUST00000110985.2 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chr19_-_32443978 | 0.71 |
ENSMUST00000078034.5
|
Rpl9-ps6
|
ribosomal protein L9, pseudogene 6 |
| chr13_-_100912308 | 0.69 |
ENSMUST00000075550.4
|
Cenph
|
centromere protein H |
| chr3_-_107838895 | 0.69 |
ENSMUST00000133947.9
ENSMUST00000124215.2 ENSMUST00000106688.8 ENSMUST00000106687.9 |
Gstm7
|
glutathione S-transferase, mu 7 |
| chr19_+_10582987 | 0.68 |
ENSMUST00000237337.2
|
Ddb1
|
damage specific DNA binding protein 1 |
| chrX_+_149377416 | 0.68 |
ENSMUST00000112713.3
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr8_-_85696369 | 0.68 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr1_-_65225572 | 0.66 |
ENSMUST00000188109.7
|
Idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr19_-_10181243 | 0.65 |
ENSMUST00000142241.2
ENSMUST00000116542.9 ENSMUST00000025651.6 ENSMUST00000156291.2 |
Fen1
|
flap structure specific endonuclease 1 |
| chr13_+_55862437 | 0.65 |
ENSMUST00000021959.11
|
Txndc15
|
thioredoxin domain containing 15 |
| chrM_+_8603 | 0.64 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr1_+_181952302 | 0.62 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr17_-_34962823 | 0.62 |
ENSMUST00000069507.9
|
C4b
|
complement component 4B (Chido blood group) |
| chr9_+_108911547 | 0.62 |
ENSMUST00000026735.9
|
Ccdc51
|
coiled-coil domain containing 51 |
| chr7_+_101546059 | 0.61 |
ENSMUST00000143835.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr4_+_108336164 | 0.60 |
ENSMUST00000155068.2
|
Tut4
|
terminal uridylyl transferase 4 |
| chr7_-_112968533 | 0.59 |
ENSMUST00000047091.14
ENSMUST00000119278.8 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chr12_+_28699598 | 0.58 |
ENSMUST00000020959.9
|
Rnaseh1
|
ribonuclease H1 |
| chr19_-_24202344 | 0.57 |
ENSMUST00000099558.5
ENSMUST00000232956.2 |
Tjp2
|
tight junction protein 2 |
| chr1_+_52158721 | 0.56 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr8_+_34621717 | 0.56 |
ENSMUST00000239436.2
ENSMUST00000033933.8 |
Saraf
|
store-operated calcium entry-associated regulatory factor |
| chr6_+_17693941 | 0.55 |
ENSMUST00000115420.8
ENSMUST00000115419.8 |
St7
|
suppression of tumorigenicity 7 |
| chr11_-_106205320 | 0.54 |
ENSMUST00000167143.2
|
Cd79b
|
CD79B antigen |
| chr2_-_29677634 | 0.54 |
ENSMUST00000177467.8
ENSMUST00000113807.10 |
Trub2
|
TruB pseudouridine (psi) synthase family member 2 |
| chr19_+_44551280 | 0.54 |
ENSMUST00000040455.5
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr12_+_83567240 | 0.54 |
ENSMUST00000021645.9
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr15_+_102427149 | 0.53 |
ENSMUST00000146756.8
ENSMUST00000142194.3 |
Tarbp2
|
TARBP2, RISC loading complex RNA binding subunit |
| chr12_-_86773160 | 0.53 |
ENSMUST00000021682.9
|
Angel1
|
angel homolog 1 |
| chr3_-_61273228 | 0.53 |
ENSMUST00000066298.3
|
B430305J03Rik
|
RIKEN cDNA B430305J03 gene |
| chr7_-_19449319 | 0.53 |
ENSMUST00000032555.10
ENSMUST00000093552.12 |
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr1_+_52158693 | 0.50 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr7_-_46558754 | 0.50 |
ENSMUST00000209538.2
|
Tsg101
|
tumor susceptibility gene 101 |
| chr4_+_136197066 | 0.49 |
ENSMUST00000170102.8
ENSMUST00000105849.9 ENSMUST00000129230.3 |
Luzp1
|
leucine zipper protein 1 |
| chr15_+_81900570 | 0.49 |
ENSMUST00000069530.13
ENSMUST00000168581.8 ENSMUST00000164779.2 |
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr3_-_86906591 | 0.49 |
ENSMUST00000063869.11
ENSMUST00000029717.4 |
Cd1d1
|
CD1d1 antigen |
| chr3_+_109481223 | 0.49 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr7_-_55669702 | 0.48 |
ENSMUST00000052204.6
|
Nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) |
| chr2_-_34760960 | 0.48 |
ENSMUST00000028225.12
|
Psmd5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr17_+_34129221 | 0.47 |
ENSMUST00000174541.9
|
Daxx
|
Fas death domain-associated protein |
| chrM_+_7758 | 0.47 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr2_-_134486039 | 0.46 |
ENSMUST00000038228.11
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr11_+_80320558 | 0.46 |
ENSMUST00000173565.2
|
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr8_-_61407760 | 0.45 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr6_-_120334400 | 0.44 |
ENSMUST00000112703.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr7_-_16761732 | 0.44 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr5_-_5564730 | 0.42 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr2_+_36026395 | 0.42 |
ENSMUST00000028250.9
|
Mrrf
|
mitochondrial ribosome recycling factor |
| chr9_-_96246460 | 0.40 |
ENSMUST00000034983.7
|
Atp1b3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr12_+_83567303 | 0.40 |
ENSMUST00000222502.2
|
Dcaf4
|
DDB1 and CUL4 associated factor 4 |
| chr17_+_71511642 | 0.40 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr1_+_179788675 | 0.40 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr6_-_108162513 | 0.40 |
ENSMUST00000167338.8
ENSMUST00000172188.2 ENSMUST00000032191.16 |
Sumf1
|
sulfatase modifying factor 1 |
| chr15_+_36174156 | 0.39 |
ENSMUST00000180159.8
ENSMUST00000057177.7 |
Polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr6_-_122778598 | 0.39 |
ENSMUST00000165884.8
|
Slc2a3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr5_+_90638580 | 0.39 |
ENSMUST00000042755.7
ENSMUST00000200693.2 |
Afp
|
alpha fetoprotein |
| chr13_+_93441447 | 0.38 |
ENSMUST00000109497.8
ENSMUST00000109498.8 ENSMUST00000060490.11 ENSMUST00000109492.9 ENSMUST00000109496.8 ENSMUST00000109495.8 |
Homer1
|
homer scaffolding protein 1 |
| chr15_+_79578141 | 0.38 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr14_-_52248324 | 0.38 |
ENSMUST00000226964.2
|
Zfp219
|
zinc finger protein 219 |
| chr16_-_22676264 | 0.38 |
ENSMUST00000232075.2
ENSMUST00000004576.8 |
Tbccd1
|
TBCC domain containing 1 |
| chrM_+_9459 | 0.37 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_+_10874498 | 0.37 |
ENSMUST00000080517.14
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chr11_+_6241660 | 0.37 |
ENSMUST00000101554.9
ENSMUST00000093350.10 |
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr7_+_101545547 | 0.36 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr2_-_155424576 | 0.36 |
ENSMUST00000126322.8
|
Gss
|
glutathione synthetase |
| chr6_+_115578863 | 0.36 |
ENSMUST00000000449.9
|
Mkrn2
|
makorin, ring finger protein, 2 |
| chr8_+_47070461 | 0.35 |
ENSMUST00000210534.2
|
Casp3
|
caspase 3 |
| chrM_+_7779 | 0.35 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr6_-_120334382 | 0.34 |
ENSMUST00000032283.12
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr8_+_85598734 | 0.34 |
ENSMUST00000170296.2
ENSMUST00000136026.8 |
Syce2
|
synaptonemal complex central element protein 2 |
| chr18_+_36926929 | 0.33 |
ENSMUST00000001419.10
|
Zmat2
|
zinc finger, matrin type 2 |
| chr19_+_36061096 | 0.33 |
ENSMUST00000025714.9
|
Rpp30
|
ribonuclease P/MRP 30 subunit |
| chr10_-_128759817 | 0.33 |
ENSMUST00000131271.2
|
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr1_-_171108754 | 0.32 |
ENSMUST00000073120.11
|
Ppox
|
protoporphyrinogen oxidase |
| chr4_-_133600308 | 0.32 |
ENSMUST00000137486.3
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr19_+_57441332 | 0.32 |
ENSMUST00000026073.14
ENSMUST00000026072.5 ENSMUST00000238107.2 |
Trub1
|
TruB pseudouridine (psi) synthase family member 1 |
| chr11_-_84710276 | 0.31 |
ENSMUST00000018549.8
|
Mrm1
|
mitochondrial rRNA methyltransferase 1 |
| chr3_+_89970088 | 0.30 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chrX_+_149372903 | 0.30 |
ENSMUST00000080884.11
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr15_+_76264695 | 0.30 |
ENSMUST00000096385.11
ENSMUST00000160728.8 |
Mroh1
|
maestro heat-like repeat family member 1 |
| chr8_+_25507227 | 0.29 |
ENSMUST00000033961.7
ENSMUST00000210536.2 |
Tm2d2
|
TM2 domain containing 2 |
| chr8_+_70583971 | 0.29 |
ENSMUST00000211898.2
ENSMUST00000095273.7 |
Nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr11_+_97594220 | 0.28 |
ENSMUST00000103147.5
|
Psmb3
|
proteasome (prosome, macropain) subunit, beta type 3 |
| chr5_-_123127346 | 0.28 |
ENSMUST00000118027.8
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr3_-_137837117 | 0.27 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr7_+_25016492 | 0.27 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr4_+_115641996 | 0.27 |
ENSMUST00000177280.8
ENSMUST00000176047.8 |
Atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr4_+_10874537 | 0.27 |
ENSMUST00000101504.3
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chrX_+_73352694 | 0.26 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr7_-_101749433 | 0.26 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr9_+_96140781 | 0.26 |
ENSMUST00000190104.7
ENSMUST00000179416.8 ENSMUST00000189606.7 |
Tfdp2
|
transcription factor Dp 2 |
| chr3_-_51184730 | 0.26 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr17_-_71160477 | 0.25 |
ENSMUST00000118283.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr9_+_8544143 | 0.24 |
ENSMUST00000050433.8
ENSMUST00000217462.2 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr11_-_96807233 | 0.24 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr14_-_78970160 | 0.24 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr16_-_84632513 | 0.24 |
ENSMUST00000023608.14
|
Atp5j
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F |
| chr6_-_148846247 | 0.24 |
ENSMUST00000111562.8
ENSMUST00000081956.12 |
Sinhcaf
|
SIN3-HDAC complex associated factor |
| chr3_-_51184895 | 0.24 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr13_+_25127127 | 0.23 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr6_-_120334302 | 0.23 |
ENSMUST00000163827.8
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr16_+_3854806 | 0.23 |
ENSMUST00000137748.8
ENSMUST00000006136.11 ENSMUST00000157044.8 ENSMUST00000120009.8 |
Dnase1
|
deoxyribonuclease I |
| chr9_+_43996236 | 0.22 |
ENSMUST00000065461.9
ENSMUST00000176416.8 |
Usp2
|
ubiquitin specific peptidase 2 |
| chr12_-_69205882 | 0.22 |
ENSMUST00000037023.9
|
Rps29
|
ribosomal protein S29 |
| chr17_-_33904345 | 0.21 |
ENSMUST00000234474.2
ENSMUST00000139302.8 ENSMUST00000114385.9 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr11_-_96807273 | 0.21 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr1_-_169358912 | 0.21 |
ENSMUST00000192248.2
ENSMUST00000028000.13 |
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr11_+_115671523 | 0.19 |
ENSMUST00000239299.2
|
Tmem94
|
transmembrane protein 94 |
| chr8_+_12965876 | 0.19 |
ENSMUST00000110876.9
ENSMUST00000110879.9 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr10_-_118705029 | 0.19 |
ENSMUST00000004281.10
|
Dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr17_-_25459086 | 0.19 |
ENSMUST00000038973.7
ENSMUST00000115154.11 |
Gnptg
|
N-acetylglucosamine-1-phosphotransferase, gamma subunit |
| chr3_-_90340910 | 0.19 |
ENSMUST00000196530.2
|
Ints3
|
integrator complex subunit 3 |
| chr13_+_100788087 | 0.19 |
ENSMUST00000190165.7
ENSMUST00000185767.7 |
Taf9
|
TATA-box binding protein associated factor 9 |
| chr10_-_126956991 | 0.18 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr11_+_93776650 | 0.18 |
ENSMUST00000107853.8
ENSMUST00000107850.8 |
Mbtd1
|
mbt domain containing 1 |
| chr19_+_53298906 | 0.18 |
ENSMUST00000003870.15
|
Mxi1
|
MAX interactor 1, dimerization protein |
| chr2_-_152673585 | 0.18 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr17_+_36152559 | 0.18 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr7_-_16761790 | 0.18 |
ENSMUST00000003183.12
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr7_+_129193581 | 0.17 |
ENSMUST00000084519.7
|
Wdr11
|
WD repeat domain 11 |
| chr19_-_12742811 | 0.17 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr17_+_47922497 | 0.15 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr5_+_20112500 | 0.15 |
ENSMUST00000101558.10
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr16_-_59459745 | 0.15 |
ENSMUST00000099646.10
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr9_-_45115381 | 0.15 |
ENSMUST00000034599.15
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr11_-_70537878 | 0.14 |
ENSMUST00000014750.15
|
Slc25a11
|
solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 |
| chr9_+_119956755 | 0.14 |
ENSMUST00000035105.7
ENSMUST00000217317.2 |
Rpsa
|
ribosomal protein SA |
| chr17_-_33904386 | 0.14 |
ENSMUST00000087582.13
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr19_-_6191440 | 0.13 |
ENSMUST00000025893.7
|
Arl2
|
ADP-ribosylation factor-like 2 |
| chr15_+_102392586 | 0.13 |
ENSMUST00000229061.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr17_-_34247016 | 0.13 |
ENSMUST00000236627.2
ENSMUST00000237759.2 ENSMUST00000045467.14 ENSMUST00000114303.4 |
H2-Ke6
|
H2-K region expressed gene 6 |
| chr17_+_41121979 | 0.12 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr9_+_96140750 | 0.12 |
ENSMUST00000186609.7
|
Tfdp2
|
transcription factor Dp 2 |
| chr3_+_87837566 | 0.12 |
ENSMUST00000055984.7
|
Isg20l2
|
interferon stimulated exonuclease gene 20-like 2 |
| chr13_-_113317431 | 0.12 |
ENSMUST00000038212.14
|
Gzmk
|
granzyme K |
| chr4_+_98919183 | 0.11 |
ENSMUST00000030280.7
|
Angptl3
|
angiopoietin-like 3 |
| chr5_-_5564873 | 0.11 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr11_+_93776965 | 0.11 |
ENSMUST00000063718.11
ENSMUST00000107854.9 |
Mbtd1
|
mbt domain containing 1 |
| chr10_+_86599836 | 0.11 |
ENSMUST00000218802.2
|
Gm49358
|
predicted gene, 49358 |
| chr7_-_29772226 | 0.09 |
ENSMUST00000183115.8
ENSMUST00000182919.8 ENSMUST00000183190.2 ENSMUST00000080834.15 |
Zfp82
|
zinc finger protein 82 |
| chr7_-_80597246 | 0.09 |
ENSMUST00000120285.8
|
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr7_-_101749140 | 0.09 |
ENSMUST00000106934.2
|
Art5
|
ADP-ribosyltransferase 5 |
| chr15_-_81900335 | 0.09 |
ENSMUST00000152227.8
|
Desi1
|
desumoylating isopeptidase 1 |
| chr16_+_44215136 | 0.08 |
ENSMUST00000099742.9
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr1_-_170695328 | 0.08 |
ENSMUST00000027974.7
|
Atf6
|
activating transcription factor 6 |
| chrX_+_41591410 | 0.08 |
ENSMUST00000005839.11
|
Sh2d1a
|
SH2 domain containing 1A |
| chr3_+_20011405 | 0.08 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr19_+_10181378 | 0.08 |
ENSMUST00000040372.14
|
Tmem258
|
transmembrane protein 258 |
| chrX_-_100266032 | 0.07 |
ENSMUST00000120389.8
ENSMUST00000156473.8 ENSMUST00000077876.4 |
Snx12
|
sorting nexin 12 |
| chr3_+_20011251 | 0.07 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr13_+_55517545 | 0.07 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr6_-_131667026 | 0.07 |
ENSMUST00000080619.3
|
Tas2r114
|
taste receptor, type 2, member 114 |
| chrX_+_41591476 | 0.07 |
ENSMUST00000115070.8
ENSMUST00000153948.2 |
Sh2d1a
|
SH2 domain containing 1A |
| chr5_-_87240405 | 0.07 |
ENSMUST00000132667.2
ENSMUST00000145617.8 ENSMUST00000094649.11 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr3_+_5283606 | 0.07 |
ENSMUST00000026284.13
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr15_-_96917804 | 0.07 |
ENSMUST00000231039.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr7_-_127048165 | 0.07 |
ENSMUST00000106299.2
|
Zfp689
|
zinc finger protein 689 |
| chr2_+_29533799 | 0.06 |
ENSMUST00000238899.2
|
Rapgef1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr5_-_148988110 | 0.06 |
ENSMUST00000110505.8
|
Hmgb1
|
high mobility group box 1 |
| chr5_-_72800070 | 0.06 |
ENSMUST00000087213.12
|
Cnga1
|
cyclic nucleotide gated channel alpha 1 |
| chr3_+_5283577 | 0.06 |
ENSMUST00000175866.8
|
Zfhx4
|
zinc finger homeodomain 4 |
| chr17_+_43671314 | 0.06 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr2_-_86640362 | 0.06 |
ENSMUST00000216117.2
|
Olfr141
|
olfactory receptor 141 |
| chr16_-_58930996 | 0.05 |
ENSMUST00000076262.4
|
Olfr193
|
olfactory receptor 193 |
| chr11_-_99695272 | 0.05 |
ENSMUST00000105056.2
|
Gm11554
|
predicted gene 11554 |
| chr18_+_65183987 | 0.05 |
ENSMUST00000236103.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr10_-_41592630 | 0.05 |
ENSMUST00000189488.3
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr4_+_109092459 | 0.05 |
ENSMUST00000106631.9
|
Calr4
|
calreticulin 4 |
| chr5_-_135092904 | 0.05 |
ENSMUST00000111205.8
ENSMUST00000141309.2 |
Bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr5_+_20112704 | 0.04 |
ENSMUST00000115267.7
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_-_61283911 | 0.04 |
ENSMUST00000111234.10
ENSMUST00000224371.2 |
Tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr7_-_19856504 | 0.04 |
ENSMUST00000079099.2
|
Vmn1r240
|
vomeronasal 1 receptor 240 |
| chr8_+_55024446 | 0.04 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr17_+_14087827 | 0.04 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr1_-_46893206 | 0.04 |
ENSMUST00000027131.6
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chrX_+_134894573 | 0.03 |
ENSMUST00000058119.9
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr15_-_96918203 | 0.03 |
ENSMUST00000166223.2
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr4_-_118667141 | 0.03 |
ENSMUST00000084313.5
ENSMUST00000219094.2 |
Olfr1335
|
olfactory receptor 1335 |
| chr7_+_51528715 | 0.03 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr14_-_20706556 | 0.03 |
ENSMUST00000090469.8
|
Myoz1
|
myozenin 1 |
| chr2_-_89313477 | 0.03 |
ENSMUST00000099778.2
|
Olfr1241
|
olfactory receptor 1241 |
| chr7_+_23702516 | 0.03 |
ENSMUST00000173571.2
|
V1rd19
|
vomeronasal 1 receptor, D19 |
| chr6_+_14901343 | 0.03 |
ENSMUST00000115477.8
|
Foxp2
|
forkhead box P2 |
| chr19_+_56276343 | 0.03 |
ENSMUST00000095948.11
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr6_+_57086439 | 0.03 |
ENSMUST00000228270.2
|
Vmn1r10
|
vomeronasal 1 receptor 10 |
| chr2_+_85519775 | 0.03 |
ENSMUST00000054868.2
|
Olfr1008
|
olfactory receptor 1008 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Brca1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.4 | 1.4 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.4 | 1.1 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.3 | 1.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 1.0 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.2 | 1.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.2 | 0.5 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 0.5 | GO:0048003 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.2 | 0.5 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 1.8 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 0.5 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.7 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.7 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.3 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.5 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 0.5 | GO:0050689 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.6 | GO:0045900 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.6 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.4 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 1.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.2 | GO:0010982 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.0 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) response to cycloheximide(GO:0046898) |
| 0.0 | 0.2 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.8 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.0 | 0.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.0 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0015870 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 2.4 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 2.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 0.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.1 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.5 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.9 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.6 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.5 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.3 | 1.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 1.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.3 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.1 | 0.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 1.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.3 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.4 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.6 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 3.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.5 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) NADH binding(GO:0070404) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID EPO PATHWAY | EPO signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.1 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.2 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |