Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
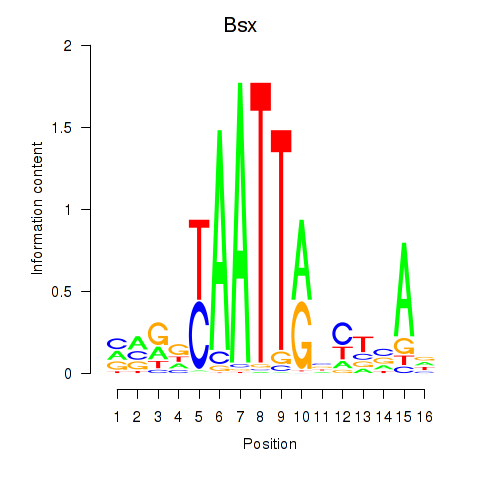
Results for Bsx
Z-value: 0.73
Transcription factors associated with Bsx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bsx
|
ENSMUSG00000054360.5 | brain specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bsx | mm39_v1_chr9_+_40785277_40785423 | -0.16 | 8.0e-01 | Click! |
Activity profile of Bsx motif
Sorted Z-values of Bsx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_23929490 | 1.10 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3 |
| chr3_+_54268523 | 0.72 |
ENSMUST00000117373.8
ENSMUST00000107985.10 ENSMUST00000073012.13 ENSMUST00000081564.13 |
Postn
|
periostin, osteoblast specific factor |
| chr13_-_21900313 | 0.66 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr13_+_21900554 | 0.39 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr11_-_109502243 | 0.33 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr5_-_23881353 | 0.33 |
ENSMUST00000198661.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr2_-_32277773 | 0.23 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr15_-_8740218 | 0.23 |
ENSMUST00000005493.14
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr5_-_137015683 | 0.21 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr15_+_78810919 | 0.20 |
ENSMUST00000089377.6
|
Lgals1
|
lectin, galactose binding, soluble 1 |
| chrM_+_7758 | 0.19 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr3_+_89153258 | 0.18 |
ENSMUST00000040888.12
|
Krtcap2
|
keratinocyte associated protein 2 |
| chrM_+_7779 | 0.17 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr4_+_8690398 | 0.17 |
ENSMUST00000127476.8
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr11_+_106916430 | 0.16 |
ENSMUST00000140447.4
|
1810010H24Rik
|
RIKEN cDNA 1810010H24 gene |
| chr7_+_3648264 | 0.14 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr11_+_119989736 | 0.13 |
ENSMUST00000106223.4
ENSMUST00000185558.2 |
Ndufaf8
|
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
| chr4_+_28813125 | 0.13 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr8_+_57964921 | 0.12 |
ENSMUST00000067925.8
|
Hmgb2
|
high mobility group box 2 |
| chr5_+_138185747 | 0.12 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr9_-_21003268 | 0.12 |
ENSMUST00000115487.3
|
Raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr12_-_99529767 | 0.11 |
ENSMUST00000176928.3
ENSMUST00000223484.2 |
Foxn3
|
forkhead box N3 |
| chr16_+_51851948 | 0.10 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr16_+_51851917 | 0.10 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr4_+_28813152 | 0.09 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chr2_+_110427643 | 0.09 |
ENSMUST00000045972.13
ENSMUST00000111026.3 |
Slc5a12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12 |
| chr3_-_15640045 | 0.08 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr15_+_25774070 | 0.08 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr8_+_57964956 | 0.08 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr5_+_64969679 | 0.08 |
ENSMUST00000166409.6
ENSMUST00000197879.2 |
Klf3
|
Kruppel-like factor 3 (basic) |
| chr3_+_90201388 | 0.08 |
ENSMUST00000199607.5
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr9_-_111086528 | 0.08 |
ENSMUST00000199404.2
|
Mlh1
|
mutL homolog 1 |
| chrM_+_8603 | 0.07 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr18_+_36877709 | 0.07 |
ENSMUST00000007042.6
ENSMUST00000237095.2 |
Ik
|
IK cytokine |
| chr6_-_116084810 | 0.07 |
ENSMUST00000204353.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr4_+_136038243 | 0.07 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr3_-_15902583 | 0.06 |
ENSMUST00000108354.8
ENSMUST00000108349.2 ENSMUST00000108352.9 ENSMUST00000108350.8 ENSMUST00000050623.11 |
Sirpb1c
|
signal-regulatory protein beta 1C |
| chr2_-_12306722 | 0.06 |
ENSMUST00000028106.11
|
Itga8
|
integrin alpha 8 |
| chr4_+_136038301 | 0.06 |
ENSMUST00000084219.12
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr5_+_144037171 | 0.06 |
ENSMUST00000041804.8
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr7_+_78922947 | 0.06 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr6_+_72074545 | 0.06 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr5_+_134961205 | 0.06 |
ENSMUST00000047196.14
ENSMUST00000111221.9 ENSMUST00000111219.8 ENSMUST00000068617.12 |
Mettl27
|
methyltransferase like 27 |
| chr11_+_116734104 | 0.05 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr16_+_51851588 | 0.05 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr8_-_68270870 | 0.05 |
ENSMUST00000059374.5
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_137015873 | 0.05 |
ENSMUST00000004968.11
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr7_+_131161951 | 0.05 |
ENSMUST00000084502.7
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr7_+_78432867 | 0.05 |
ENSMUST00000032840.5
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr7_+_131162439 | 0.05 |
ENSMUST00000207442.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr4_+_136038263 | 0.04 |
ENSMUST00000105850.8
ENSMUST00000148843.10 |
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr5_-_104125226 | 0.04 |
ENSMUST00000048118.15
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr1_+_82817794 | 0.04 |
ENSMUST00000186043.2
|
Agfg1
|
ArfGAP with FG repeats 1 |
| chr12_+_103498542 | 0.04 |
ENSMUST00000021631.12
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr2_-_32278245 | 0.04 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr3_-_89153135 | 0.03 |
ENSMUST00000041022.15
|
Trim46
|
tripartite motif-containing 46 |
| chr1_-_173018204 | 0.03 |
ENSMUST00000215878.2
ENSMUST00000201132.3 |
Olfr1406
|
olfactory receptor 1406 |
| chr3_-_16060491 | 0.03 |
ENSMUST00000108347.3
|
Gm5150
|
predicted gene 5150 |
| chr9_-_50657800 | 0.03 |
ENSMUST00000239417.2
ENSMUST00000034564.4 |
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr12_+_100076407 | 0.03 |
ENSMUST00000021595.10
|
Psmc1
|
protease (prosome, macropain) 26S subunit, ATPase 1 |
| chr2_-_85632888 | 0.03 |
ENSMUST00000217410.3
ENSMUST00000216425.3 |
Olfr1016
|
olfactory receptor 1016 |
| chr1_-_191129223 | 0.03 |
ENSMUST00000067976.9
|
Ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr17_-_31496352 | 0.03 |
ENSMUST00000024832.9
|
Rsph1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr1_-_90897329 | 0.03 |
ENSMUST00000130042.2
ENSMUST00000027529.12 |
Rab17
|
RAB17, member RAS oncogene family |
| chr7_+_126808016 | 0.02 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr4_-_14621805 | 0.02 |
ENSMUST00000042221.14
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr3_-_64197130 | 0.02 |
ENSMUST00000233531.2
ENSMUST00000176328.9 |
Vmn2r3
|
vomeronasal 2, receptor 3 |
| chr1_+_40478926 | 0.02 |
ENSMUST00000173514.8
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr3_+_55689921 | 0.02 |
ENSMUST00000075422.6
|
Mab21l1
|
mab-21-like 1 |
| chr10_-_6930376 | 0.02 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr2_+_85868891 | 0.02 |
ENSMUST00000218397.2
|
Olfr1033
|
olfactory receptor 1033 |
| chr6_-_144993451 | 0.02 |
ENSMUST00000123930.8
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr2_-_85245221 | 0.02 |
ENSMUST00000099926.2
|
Olfr993
|
olfactory receptor 993 |
| chr7_-_119058489 | 0.02 |
ENSMUST00000207887.3
ENSMUST00000239424.2 ENSMUST00000033255.8 |
Gp2
|
glycoprotein 2 (zymogen granule membrane) |
| chr16_-_63684477 | 0.02 |
ENSMUST00000232654.2
ENSMUST00000064405.8 |
Epha3
|
Eph receptor A3 |
| chr2_-_113659360 | 0.02 |
ENSMUST00000024005.8
|
Scg5
|
secretogranin V |
| chr11_+_73827503 | 0.02 |
ENSMUST00000214210.2
|
Olfr23
|
olfactory receptor 23 |
| chr12_+_36042899 | 0.02 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr7_+_103628383 | 0.02 |
ENSMUST00000098185.2
|
Olfr635
|
olfactory receptor 635 |
| chr13_-_21847556 | 0.02 |
ENSMUST00000214321.2
|
Olfr1361
|
olfactory receptor 1361 |
| chr7_+_101545547 | 0.02 |
ENSMUST00000035395.14
ENSMUST00000106973.8 ENSMUST00000144207.9 |
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr11_-_99412084 | 0.01 |
ENSMUST00000076948.2
|
Krt39
|
keratin 39 |
| chr3_-_58729732 | 0.01 |
ENSMUST00000191233.4
|
Mindy4b-ps
|
MINDY lysine 48 deubiquitinase 4B, pseudogene |
| chr16_-_58932235 | 0.01 |
ENSMUST00000207935.2
ENSMUST00000208455.3 |
Olfr193
|
olfactory receptor 193 |
| chr17_-_48144094 | 0.01 |
ENSMUST00000131971.2
ENSMUST00000129360.2 ENSMUST00000113280.8 ENSMUST00000132125.8 |
Mdfi
|
MyoD family inhibitor |
| chr11_-_99412162 | 0.01 |
ENSMUST00000107445.8
|
Krt39
|
keratin 39 |
| chr6_+_63232955 | 0.01 |
ENSMUST00000095852.5
|
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr16_-_63684425 | 0.01 |
ENSMUST00000232049.2
|
Epha3
|
Eph receptor A3 |
| chr1_+_171041583 | 0.01 |
ENSMUST00000111328.8
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr17_-_48144181 | 0.01 |
ENSMUST00000152455.8
ENSMUST00000035375.14 |
Mdfi
|
MyoD family inhibitor |
| chr3_-_15491482 | 0.01 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr19_+_4644365 | 0.01 |
ENSMUST00000113825.4
|
Pcx
|
pyruvate carboxylase |
| chr1_-_172964941 | 0.01 |
ENSMUST00000200689.5
|
Olfr1408
|
olfactory receptor 1408 |
| chr5_-_104125270 | 0.01 |
ENSMUST00000112803.3
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr4_-_14621497 | 0.01 |
ENSMUST00000149633.2
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr1_+_130659700 | 0.01 |
ENSMUST00000039323.8
|
AA986860
|
expressed sequence AA986860 |
| chr14_+_50690511 | 0.01 |
ENSMUST00000089838.2
|
Olfr740
|
olfactory receptor 740 |
| chr1_-_163552693 | 0.01 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr5_-_104125192 | 0.01 |
ENSMUST00000120320.8
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr2_+_69050315 | 0.01 |
ENSMUST00000005364.12
ENSMUST00000112317.3 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr6_-_124756645 | 0.01 |
ENSMUST00000147669.2
ENSMUST00000128697.8 ENSMUST00000032218.10 ENSMUST00000112475.9 |
Lrrc23
|
leucine rich repeat containing 23 |
| chr13_+_27305681 | 0.01 |
ENSMUST00000080755.12
ENSMUST00000164964.4 |
Prl3d2
|
prolactin family 3, subfamily d, member 1 |
| chr14_+_64229914 | 0.01 |
ENSMUST00000058229.6
|
Rp1l1
|
retinitis pigmentosa 1 homolog like 1 |
| chr18_+_56565188 | 0.01 |
ENSMUST00000070166.6
|
Gramd3
|
GRAM domain containing 3 |
| chr7_+_102526329 | 0.01 |
ENSMUST00000098216.2
|
Olfr568
|
olfactory receptor 568 |
| chr4_-_118549953 | 0.01 |
ENSMUST00000216226.2
|
Olfr1342
|
olfactory receptor 1342 |
| chr13_+_65300040 | 0.01 |
ENSMUST00000058907.4
|
Olfr466
|
olfactory receptor 466 |
| chr16_-_59151501 | 0.01 |
ENSMUST00000213910.2
|
Olfr205
|
olfactory receptor 205 |
| chr2_-_89587193 | 0.01 |
ENSMUST00000215988.2
|
Olfr1253
|
olfactory receptor 1253 |
| chr1_+_150195158 | 0.01 |
ENSMUST00000165062.8
ENSMUST00000191228.7 ENSMUST00000186572.7 ENSMUST00000185698.2 |
Pdc
|
phosducin |
| chr17_+_93506435 | 0.01 |
ENSMUST00000234646.2
ENSMUST00000234081.2 |
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr14_-_109151590 | 0.01 |
ENSMUST00000100322.4
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chrY_+_80146479 | 0.01 |
ENSMUST00000179811.2
|
Gm21760
|
predicted gene, 21760 |
| chr1_-_4479445 | 0.01 |
ENSMUST00000208660.2
|
Rp1
|
retinitis pigmentosa 1 (human) |
| chrY_-_40270796 | 0.01 |
ENSMUST00000177713.2
|
Gm21865
|
predicted gene, 21865 |
| chr17_+_93506590 | 0.01 |
ENSMUST00000064775.8
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr14_+_65504067 | 0.01 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr5_+_90708962 | 0.01 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr6_-_56546088 | 0.01 |
ENSMUST00000203372.3
|
Pde1c
|
phosphodiesterase 1C |
| chr10_+_97482946 | 0.01 |
ENSMUST00000105285.4
|
Epyc
|
epiphycan |
| chr11_-_33942981 | 0.01 |
ENSMUST00000238903.2
|
Kcnip1
|
Kv channel-interacting protein 1 |
| chr2_+_85597442 | 0.01 |
ENSMUST00000216397.3
|
Olfr1013
|
olfactory receptor 1013 |
| chrX_+_96116202 | 0.01 |
ENSMUST00000033556.4
|
Pgr15l
|
G protein-coupled receptor 15-like |
| chr9_+_110306020 | 0.00 |
ENSMUST00000198858.5
|
Kif9
|
kinesin family member 9 |
| chr16_-_59149895 | 0.00 |
ENSMUST00000074125.3
|
Olfr205
|
olfactory receptor 205 |
| chr16_-_59166089 | 0.00 |
ENSMUST00000084791.4
|
Olfr206
|
olfactory receptor 206 |
| chr7_+_88079465 | 0.00 |
ENSMUST00000107256.4
|
Rab38
|
RAB38, member RAS oncogene family |
| chr2_-_88176836 | 0.00 |
ENSMUST00000216713.2
|
Olfr1177-ps
|
olfactory receptor 1177, pseudogene |
| chrY_+_52778041 | 0.00 |
ENSMUST00000178673.2
|
Gm21258
|
predicted gene, 21258 |
| chrY_-_35875442 | 0.00 |
ENSMUST00000180332.2
|
Gm20896
|
predicted gene, 20896 |
| chr9_-_38976247 | 0.00 |
ENSMUST00000215049.2
|
Olfr937
|
olfactory receptor 937 |
| chr4_-_91264670 | 0.00 |
ENSMUST00000107109.9
ENSMUST00000107111.9 ENSMUST00000107120.8 |
Elavl2
|
ELAV like RNA binding protein 1 |
| chr7_-_98305737 | 0.00 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr7_+_63835285 | 0.00 |
ENSMUST00000206263.2
ENSMUST00000206107.2 ENSMUST00000205731.2 ENSMUST00000206706.2 ENSMUST00000205690.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr4_-_91264727 | 0.00 |
ENSMUST00000107124.10
|
Elavl2
|
ELAV like RNA binding protein 1 |
| chr14_-_36820304 | 0.00 |
ENSMUST00000022337.11
|
Cdhr1
|
cadherin-related family member 1 |
| chr7_+_107700056 | 0.00 |
ENSMUST00000215159.3
|
Olfr483
|
olfactory receptor 483 |
| chr9_+_39128655 | 0.00 |
ENSMUST00000079650.2
|
Olfr944
|
olfactory receptor 944 |
| chr2_+_110551976 | 0.00 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
| chr7_+_63835154 | 0.00 |
ENSMUST00000177102.8
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr2_-_86982968 | 0.00 |
ENSMUST00000216378.2
|
Olfr1111
|
olfactory receptor 1111 |
| chr7_-_102143980 | 0.00 |
ENSMUST00000058750.4
|
Olfr545
|
olfactory receptor 545 |
| chr2_+_89642395 | 0.00 |
ENSMUST00000214508.2
|
Olfr1255
|
olfactory receptor 1255 |
| chr3_+_138019040 | 0.00 |
ENSMUST00000013455.13
ENSMUST00000106247.2 |
Adh6a
|
alcohol dehydrogenase 6A (class V) |
| chr18_+_37433852 | 0.00 |
ENSMUST00000051754.2
|
Pcdhb3
|
protocadherin beta 3 |
| chr6_-_144993362 | 0.00 |
ENSMUST00000149769.6
|
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr4_-_114991174 | 0.00 |
ENSMUST00000051400.8
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr9_-_70841881 | 0.00 |
ENSMUST00000214995.2
|
Lipc
|
lipase, hepatic |
| chr19_-_56378309 | 0.00 |
ENSMUST00000166203.2
ENSMUST00000167239.8 |
Nrap
|
nebulin-related anchoring protein |
| chr16_-_59135768 | 0.00 |
ENSMUST00000072517.2
|
Olfr204
|
olfactory receptor 204 |
| chr3_-_113371392 | 0.00 |
ENSMUST00000067980.12
|
Amy1
|
amylase 1, salivary |
| chr15_+_92495007 | 0.00 |
ENSMUST00000035399.10
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chrY_+_55729767 | 0.00 |
ENSMUST00000177834.2
|
Gm21858
|
predicted gene, 21858 |
| chr4_-_129155185 | 0.00 |
ENSMUST00000145261.8
|
C77080
|
expressed sequence C77080 |
| chr10_-_23977810 | 0.00 |
ENSMUST00000170267.3
|
Taar8c
|
trace amine-associated receptor 8C |
| chr2_-_88559941 | 0.00 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bsx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.3 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0034118 | erythrocyte aggregation(GO:0034117) regulation of erythrocyte aggregation(GO:0034118) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.3 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.3 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |