Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Cdx2
Z-value: 0.81
Transcription factors associated with Cdx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
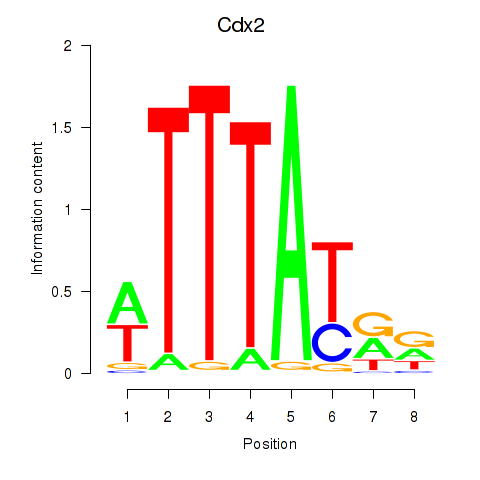
Cdx2
|
ENSMUSG00000029646.4 | caudal type homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cdx2 | mm39_v1_chr5_-_147244074_147244091 | 0.99 | 7.6e-04 | Click! |
Activity profile of Cdx2 motif
Sorted Z-values of Cdx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23765546 | 1.82 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4 |
| chr13_+_23728222 | 1.22 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr13_-_24118139 | 1.21 |
ENSMUST00000052776.4
|
H2bc1
|
H2B clustered histone 1 |
| chr13_-_22219738 | 0.79 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr2_-_59778560 | 0.38 |
ENSMUST00000153136.2
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr19_+_8828132 | 0.35 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr15_+_3300249 | 0.34 |
ENSMUST00000082424.12
ENSMUST00000159158.9 ENSMUST00000159216.10 ENSMUST00000160311.3 |
Selenop
|
selenoprotein P |
| chr14_-_54923517 | 0.28 |
ENSMUST00000125265.2
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr8_-_5155347 | 0.27 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr14_+_80237691 | 0.25 |
ENSMUST00000228749.2
ENSMUST00000088735.4 |
Olfm4
|
olfactomedin 4 |
| chr15_-_50746202 | 0.24 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr6_-_52190299 | 0.24 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308 |
| chr19_-_34856853 | 0.20 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr2_-_51039112 | 0.14 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chrX_+_72818003 | 0.14 |
ENSMUST00000002081.6
|
Srpk3
|
serine/arginine-rich protein specific kinase 3 |
| chr1_-_186438177 | 0.10 |
ENSMUST00000045288.14
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr8_+_95113066 | 0.09 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr10_-_33920300 | 0.09 |
ENSMUST00000048052.7
|
Calhm4
|
calcium homeostasis modulator family member 4 |
| chr11_-_99134885 | 0.07 |
ENSMUST00000103132.10
ENSMUST00000038214.7 |
Krt222
|
keratin 222 |
| chr12_+_113038098 | 0.07 |
ENSMUST00000012355.14
ENSMUST00000146107.8 |
Tex22
|
testis expressed gene 22 |
| chr6_-_122317484 | 0.06 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr9_+_95519654 | 0.06 |
ENSMUST00000015498.9
|
Pcolce2
|
procollagen C-endopeptidase enhancer 2 |
| chr6_+_17491231 | 0.05 |
ENSMUST00000080469.12
|
Met
|
met proto-oncogene |
| chr11_-_79394904 | 0.04 |
ENSMUST00000164465.3
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr18_-_79152504 | 0.04 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr8_-_34333573 | 0.04 |
ENSMUST00000183062.2
|
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr18_-_35841435 | 0.04 |
ENSMUST00000236738.2
ENSMUST00000237995.2 |
Dnajc18
|
DnaJ heat shock protein family (Hsp40) member C18 |
| chr1_-_179572765 | 0.04 |
ENSMUST00000211943.3
ENSMUST00000131716.4 ENSMUST00000221136.2 |
Kif28
|
kinesin family member 28 |
| chr13_-_63712140 | 0.03 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1 |
| chr2_-_25982160 | 0.03 |
ENSMUST00000114159.9
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing |
| chr3_+_13536696 | 0.03 |
ENSMUST00000191806.3
ENSMUST00000193117.3 |
Ralyl
|
RALY RNA binding protein-like |
| chr4_-_109059414 | 0.03 |
ENSMUST00000160774.8
ENSMUST00000194478.6 ENSMUST00000030288.14 ENSMUST00000162787.9 |
Osbpl9
|
oxysterol binding protein-like 9 |
| chr9_-_15023396 | 0.03 |
ENSMUST00000159985.2
|
Hephl1
|
hephaestin-like 1 |
| chr5_+_13448833 | 0.02 |
ENSMUST00000137798.10
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr6_-_71239216 | 0.02 |
ENSMUST00000129630.3
ENSMUST00000114186.9 ENSMUST00000074301.10 |
Smyd1
|
SET and MYND domain containing 1 |
| chrX_+_138585728 | 0.02 |
ENSMUST00000096313.4
|
Tbc1d8b
|
TBC1 domain family, member 8B |
| chr18_+_65715460 | 0.02 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chr18_+_65715156 | 0.02 |
ENSMUST00000237553.2
ENSMUST00000237712.2 ENSMUST00000182684.8 |
Zfp532
|
zinc finger protein 532 |
| chr3_+_126390951 | 0.02 |
ENSMUST00000171289.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr17_-_90395771 | 0.02 |
ENSMUST00000197268.5
ENSMUST00000173917.8 |
Nrxn1
|
neurexin I |
| chr10_-_33920289 | 0.02 |
ENSMUST00000218239.2
|
Calhm4
|
calcium homeostasis modulator family member 4 |
| chr14_+_54923655 | 0.01 |
ENSMUST00000038539.8
ENSMUST00000228027.2 |
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr5_+_128677863 | 0.01 |
ENSMUST00000117102.4
|
Fzd10
|
frizzled class receptor 10 |
| chr2_+_74552322 | 0.01 |
ENSMUST00000047904.4
|
Hoxd4
|
homeobox D4 |
| chr10_+_118040729 | 0.01 |
ENSMUST00000096691.5
|
Il22
|
interleukin 22 |
| chr2_+_142920386 | 0.01 |
ENSMUST00000028902.3
|
Otor
|
otoraplin |
| chrX_-_50294867 | 0.01 |
ENSMUST00000114876.9
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr11_-_99328969 | 0.01 |
ENSMUST00000017743.3
|
Krt20
|
keratin 20 |
| chrX_+_21581135 | 0.01 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr1_-_186437760 | 0.01 |
ENSMUST00000195201.2
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr5_+_67055753 | 0.01 |
ENSMUST00000122812.5
ENSMUST00000117601.8 |
Limch1
|
LIM and calponin homology domains 1 |
| chr15_+_23036535 | 0.01 |
ENSMUST00000164787.8
|
Cdh18
|
cadherin 18 |
| chr9_-_65330231 | 0.01 |
ENSMUST00000065894.7
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr15_-_101441254 | 0.01 |
ENSMUST00000023720.8
|
Krt84
|
keratin 84 |
| chr3_+_3699205 | 0.00 |
ENSMUST00000108394.3
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr6_+_42263609 | 0.00 |
ENSMUST00000238845.2
ENSMUST00000031894.13 |
Clcn1
|
chloride channel, voltage-sensitive 1 |
| chr3_+_93052089 | 0.00 |
ENSMUST00000107300.7
ENSMUST00000195515.2 |
Crnn
|
cornulin |
| chr8_+_55003359 | 0.00 |
ENSMUST00000033918.4
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr18_+_65715351 | 0.00 |
ENSMUST00000235669.2
ENSMUST00000183319.8 ENSMUST00000236523.2 ENSMUST00000237886.2 |
Zfp532
|
zinc finger protein 532 |
| chr11_-_99433984 | 0.00 |
ENSMUST00000107443.8
ENSMUST00000074253.4 |
Krt40
|
keratin 40 |
| chr18_+_44249254 | 0.00 |
ENSMUST00000212114.2
|
Gm37797
|
predicted gene, 37797 |
| chr19_-_47680528 | 0.00 |
ENSMUST00000026045.14
ENSMUST00000086923.6 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr10_+_129357661 | 0.00 |
ENSMUST00000217228.2
|
Olfr791
|
olfactory receptor 791 |
| chr3_+_126391046 | 0.00 |
ENSMUST00000106401.8
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta |
| chr17_-_31348576 | 0.00 |
ENSMUST00000024827.5
|
Tff3
|
trefoil factor 3, intestinal |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cdx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:1905006 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |