Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Cebpa_Cebpg
Z-value: 1.05
Transcription factors associated with Cebpa_Cebpg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
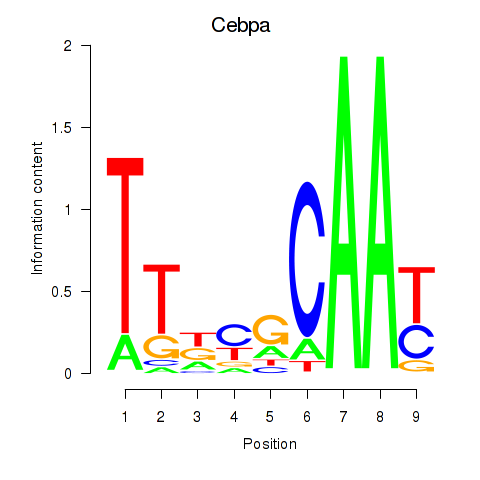
Cebpa
|
ENSMUSG00000034957.11 | CCAAT/enhancer binding protein (C/EBP), alpha |
|
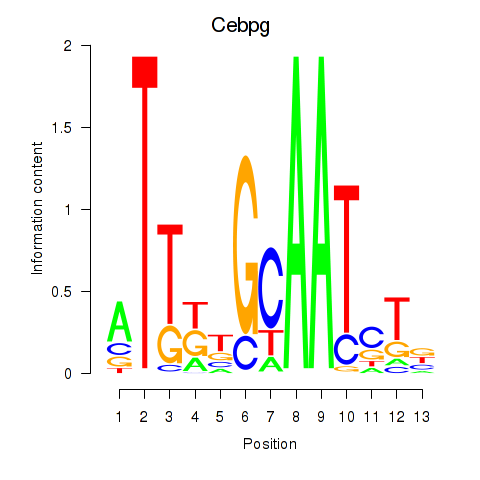
Cebpg
|
ENSMUSG00000056216.10 | CCAAT/enhancer binding protein (C/EBP), gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpg | mm39_v1_chr7_-_34755985_34756002 | -0.40 | 5.0e-01 | Click! |
| Cebpa | mm39_v1_chr7_+_34818709_34818728 | 0.27 | 6.6e-01 | Click! |
Activity profile of Cebpa_Cebpg motif
Sorted Z-values of Cebpa_Cebpg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_80765900 | 0.68 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr18_-_36859732 | 0.51 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr9_+_122752116 | 0.50 |
ENSMUST00000051667.14
|
Zfp105
|
zinc finger protein 105 |
| chr9_-_100388857 | 0.49 |
ENSMUST00000112874.4
|
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chrX_+_93679671 | 0.43 |
ENSMUST00000096368.4
|
Gspt2
|
G1 to S phase transition 2 |
| chr14_+_51181956 | 0.42 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr19_-_11618165 | 0.42 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr19_-_11618192 | 0.41 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr2_+_172235820 | 0.41 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr3_-_50398027 | 0.40 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr5_-_138169509 | 0.34 |
ENSMUST00000153867.8
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr2_-_32278245 | 0.33 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr3_+_106020545 | 0.32 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr7_-_126873219 | 0.31 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr11_-_72441054 | 0.31 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chrM_+_8603 | 0.30 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chr18_+_21077627 | 0.29 |
ENSMUST00000050004.3
|
Rnf125
|
ring finger protein 125 |
| chr4_+_150322151 | 0.28 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chr14_+_31887739 | 0.28 |
ENSMUST00000111994.10
ENSMUST00000168114.8 ENSMUST00000168034.8 |
Ncoa4
|
nuclear receptor coactivator 4 |
| chr2_-_32314017 | 0.28 |
ENSMUST00000113307.9
|
Slc25a25
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 |
| chr18_-_33346885 | 0.28 |
ENSMUST00000025236.9
|
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr1_+_40554513 | 0.27 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr8_-_79539838 | 0.27 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_+_125518736 | 0.26 |
ENSMUST00000198811.2
|
Bri3bp
|
Bri3 binding protein |
| chr15_+_102379503 | 0.25 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr6_-_3968365 | 0.25 |
ENSMUST00000031674.11
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr18_-_64649497 | 0.24 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr7_-_45083688 | 0.24 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr5_-_88823989 | 0.24 |
ENSMUST00000078945.12
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr11_+_40624763 | 0.23 |
ENSMUST00000127382.2
|
Nudcd2
|
NudC domain containing 2 |
| chr18_-_33346819 | 0.23 |
ENSMUST00000119991.8
ENSMUST00000118990.2 |
Stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr2_-_129213050 | 0.22 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr9_+_21746785 | 0.22 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chr4_-_108690741 | 0.22 |
ENSMUST00000102740.8
ENSMUST00000102741.8 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr11_-_83540175 | 0.22 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr18_-_64649620 | 0.21 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr10_+_127919142 | 0.21 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr14_+_22069709 | 0.21 |
ENSMUST00000075639.11
|
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr3_-_65299967 | 0.20 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr9_+_5298669 | 0.20 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr13_-_117162041 | 0.20 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr3_-_107893676 | 0.19 |
ENSMUST00000066530.7
ENSMUST00000012348.9 |
Gstm2
|
glutathione S-transferase, mu 2 |
| chr15_+_102379621 | 0.19 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr7_+_46496929 | 0.19 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr3_-_65300000 | 0.19 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr13_+_95460110 | 0.19 |
ENSMUST00000221807.2
|
Zbed3
|
zinc finger, BED type containing 3 |
| chr6_-_86710250 | 0.18 |
ENSMUST00000001185.14
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr7_+_127399776 | 0.18 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr10_-_117128763 | 0.18 |
ENSMUST00000092162.7
|
Lyz1
|
lysozyme 1 |
| chr10_-_111829393 | 0.18 |
ENSMUST00000161870.3
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr15_+_44482944 | 0.17 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr5_+_115417725 | 0.16 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr8_-_96615138 | 0.16 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr11_+_96932379 | 0.16 |
ENSMUST00000001485.10
|
Mrpl10
|
mitochondrial ribosomal protein L10 |
| chr5_+_35156389 | 0.16 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr19_-_3979723 | 0.16 |
ENSMUST00000051803.8
|
Aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr7_+_28136861 | 0.16 |
ENSMUST00000108292.9
ENSMUST00000108289.8 |
Gmfg
|
glia maturation factor, gamma |
| chr1_-_80255156 | 0.16 |
ENSMUST00000168372.2
|
Cul3
|
cullin 3 |
| chr17_+_28910302 | 0.15 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr4_+_130001349 | 0.15 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr1_+_143614753 | 0.15 |
ENSMUST00000145969.8
|
Glrx2
|
glutaredoxin 2 (thioltransferase) |
| chr4_-_136626073 | 0.14 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr2_+_85809620 | 0.14 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr4_-_63414188 | 0.14 |
ENSMUST00000063650.10
ENSMUST00000102867.8 ENSMUST00000107393.8 ENSMUST00000084510.8 ENSMUST00000095038.8 ENSMUST00000119294.8 ENSMUST00000095037.2 ENSMUST00000063672.10 |
Whrn
|
whirlin |
| chr1_+_169483062 | 0.14 |
ENSMUST00000027997.9
ENSMUST00000152809.3 |
Rgs5
|
regulator of G-protein signaling 5 |
| chr7_-_45084012 | 0.14 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr7_+_101530524 | 0.14 |
ENSMUST00000106978.8
|
Anapc15
|
anaphase promoting complex C subunit 15 |
| chr4_+_152123772 | 0.13 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chrX_+_108240356 | 0.13 |
ENSMUST00000139259.2
ENSMUST00000060013.4 |
Gm6377
|
predicted gene 6377 |
| chr9_+_94551929 | 0.13 |
ENSMUST00000033463.10
|
Slc9a9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9 |
| chr11_+_49327451 | 0.13 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr16_-_33916354 | 0.13 |
ENSMUST00000114973.9
ENSMUST00000232157.2 ENSMUST00000114964.8 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr9_+_107957640 | 0.13 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_+_40624466 | 0.12 |
ENSMUST00000020578.11
|
Nudcd2
|
NudC domain containing 2 |
| chr3_+_20011251 | 0.12 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr16_-_16942970 | 0.12 |
ENSMUST00000093336.8
ENSMUST00000231681.2 |
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr5_+_102872838 | 0.12 |
ENSMUST00000112853.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr19_-_12742811 | 0.12 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr2_-_20948230 | 0.12 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr15_-_55770118 | 0.12 |
ENSMUST00000110200.3
|
Sntb1
|
syntrophin, basic 1 |
| chr3_+_20011405 | 0.12 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr5_-_124003553 | 0.11 |
ENSMUST00000057145.7
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr1_+_74414354 | 0.11 |
ENSMUST00000187516.7
ENSMUST00000027368.6 |
Slc11a1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 |
| chr1_-_79838897 | 0.11 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr7_-_45109262 | 0.11 |
ENSMUST00000094434.13
|
Ftl1
|
ferritin light polypeptide 1 |
| chr1_-_170002444 | 0.11 |
ENSMUST00000111351.10
ENSMUST00000027981.8 |
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr4_-_130253703 | 0.11 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr11_+_77928736 | 0.11 |
ENSMUST00000072289.12
ENSMUST00000100784.9 ENSMUST00000073660.7 |
Flot2
|
flotillin 2 |
| chr5_-_100867520 | 0.11 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr7_+_30475819 | 0.11 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr7_-_45109075 | 0.11 |
ENSMUST00000210864.2
|
Ftl1
|
ferritin light polypeptide 1 |
| chr16_+_19578945 | 0.10 |
ENSMUST00000121344.8
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr10_+_23727325 | 0.10 |
ENSMUST00000020190.8
|
Vnn3
|
vanin 3 |
| chr7_+_27147403 | 0.10 |
ENSMUST00000037399.16
ENSMUST00000108358.8 |
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr1_-_170002526 | 0.10 |
ENSMUST00000111350.10
|
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr9_+_107957621 | 0.10 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr14_-_78866714 | 0.10 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr9_+_78522783 | 0.10 |
ENSMUST00000093812.5
|
Cd109
|
CD109 antigen |
| chr10_-_88192852 | 0.10 |
ENSMUST00000020249.2
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr11_+_106107752 | 0.10 |
ENSMUST00000021046.6
|
Ddx42
|
DEAD box helicase 42 |
| chr7_+_51530060 | 0.10 |
ENSMUST00000145049.2
|
Gas2
|
growth arrest specific 2 |
| chr5_-_137856280 | 0.10 |
ENSMUST00000110978.7
ENSMUST00000199387.2 ENSMUST00000196195.2 |
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr9_+_123901979 | 0.10 |
ENSMUST00000171719.8
|
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr15_+_44482667 | 0.09 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr11_+_108477903 | 0.09 |
ENSMUST00000146912.9
|
Cep112
|
centrosomal protein 112 |
| chr17_-_35454729 | 0.09 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chr7_+_45084300 | 0.09 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr3_-_63872189 | 0.09 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr14_+_51399692 | 0.09 |
ENSMUST00000075648.4
|
Rnase2b
|
ribonuclease, RNase A family, 2B (liver, eosinophil-derived neurotoxin) |
| chr6_+_30401864 | 0.09 |
ENSMUST00000068240.13
ENSMUST00000068259.10 ENSMUST00000132581.8 |
Klhdc10
|
kelch domain containing 10 |
| chr7_+_45084257 | 0.08 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr3_+_66127330 | 0.08 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr1_-_164281344 | 0.08 |
ENSMUST00000193367.2
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr13_-_117161921 | 0.08 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr13_+_51254852 | 0.08 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr17_+_28910393 | 0.08 |
ENSMUST00000124886.9
ENSMUST00000114758.9 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr10_+_13428638 | 0.08 |
ENSMUST00000019944.9
|
Adat2
|
adenosine deaminase, tRNA-specific 2 |
| chr5_-_92231517 | 0.08 |
ENSMUST00000202258.4
ENSMUST00000113127.7 |
G3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr14_+_60970355 | 0.07 |
ENSMUST00000162945.2
|
Spata13
|
spermatogenesis associated 13 |
| chr15_+_101982208 | 0.07 |
ENSMUST00000169681.3
ENSMUST00000229400.2 |
Eif4b
|
eukaryotic translation initiation factor 4B |
| chr9_+_123902143 | 0.07 |
ENSMUST00000168841.3
ENSMUST00000055918.7 |
Ccr2
|
chemokine (C-C motif) receptor 2 |
| chr10_-_5144699 | 0.07 |
ENSMUST00000215467.2
|
Syne1
|
spectrin repeat containing, nuclear envelope 1 |
| chr9_-_61821820 | 0.07 |
ENSMUST00000008036.9
|
Rplp1
|
ribosomal protein, large, P1 |
| chr14_+_52091156 | 0.07 |
ENSMUST00000169070.2
ENSMUST00000074477.7 |
Ear6
|
eosinophil-associated, ribonuclease A family, member 6 |
| chr9_+_107852733 | 0.07 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr11_-_102120953 | 0.07 |
ENSMUST00000107150.8
ENSMUST00000156337.2 ENSMUST00000107151.9 ENSMUST00000107152.9 |
Hdac5
|
histone deacetylase 5 |
| chr1_-_52271455 | 0.07 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr2_+_25313240 | 0.07 |
ENSMUST00000134259.8
ENSMUST00000100320.5 |
Fut7
|
fucosyltransferase 7 |
| chr4_-_130068902 | 0.07 |
ENSMUST00000105998.8
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr15_-_76191301 | 0.07 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr18_+_44237474 | 0.07 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr11_-_102120917 | 0.07 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr4_-_108690805 | 0.06 |
ENSMUST00000102742.11
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr12_+_100745333 | 0.06 |
ENSMUST00000110073.8
ENSMUST00000110069.8 |
Dglucy
|
D-glutamate cyclase |
| chr10_+_5543769 | 0.06 |
ENSMUST00000051809.10
|
Myct1
|
myc target 1 |
| chr3_-_63872079 | 0.06 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr6_-_28397997 | 0.06 |
ENSMUST00000035930.11
|
Zfp800
|
zinc finger protein 800 |
| chr6_-_86710220 | 0.06 |
ENSMUST00000113679.2
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr7_-_5128936 | 0.06 |
ENSMUST00000147835.4
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr11_-_84761472 | 0.06 |
ENSMUST00000018547.9
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr2_-_110781268 | 0.06 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr9_-_123546605 | 0.06 |
ENSMUST00000163207.2
|
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr2_-_52566583 | 0.06 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr11_-_84761538 | 0.06 |
ENSMUST00000170741.2
ENSMUST00000172405.8 ENSMUST00000100686.10 ENSMUST00000108081.9 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr12_+_100745314 | 0.06 |
ENSMUST00000069782.11
|
Dglucy
|
D-glutamate cyclase |
| chr7_-_85956319 | 0.05 |
ENSMUST00000055690.3
|
Olfr309
|
olfactory receptor 309 |
| chr18_+_44237577 | 0.05 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr11_-_77971186 | 0.05 |
ENSMUST00000021183.4
|
Eral1
|
Era (G-protein)-like 1 (E. coli) |
| chr15_-_103218876 | 0.05 |
ENSMUST00000079824.6
|
Gpr84
|
G protein-coupled receptor 84 |
| chrX_-_93585668 | 0.05 |
ENSMUST00000026142.8
|
Maged1
|
MAGE family member D1 |
| chr16_-_22847808 | 0.05 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr15_+_31225302 | 0.05 |
ENSMUST00000186425.7
|
Dap
|
death-associated protein |
| chr2_+_109522781 | 0.05 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr14_-_44057096 | 0.05 |
ENSMUST00000100691.4
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chr8_-_41507808 | 0.04 |
ENSMUST00000093534.11
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr17_+_36134122 | 0.04 |
ENSMUST00000001569.15
ENSMUST00000174080.8 |
Flot1
|
flotillin 1 |
| chr9_+_7571397 | 0.04 |
ENSMUST00000120900.8
ENSMUST00000151853.8 ENSMUST00000152878.3 |
Mmp27
|
matrix metallopeptidase 27 |
| chr11_+_66969119 | 0.04 |
ENSMUST00000108689.8
ENSMUST00000007301.14 ENSMUST00000165221.2 |
Myh3
|
myosin, heavy polypeptide 3, skeletal muscle, embryonic |
| chr14_+_66208613 | 0.04 |
ENSMUST00000144619.2
|
Clu
|
clusterin |
| chr12_-_65219328 | 0.04 |
ENSMUST00000124201.2
ENSMUST00000052201.9 ENSMUST00000222244.2 ENSMUST00000221296.2 |
Mis18bp1
|
MIS18 binding protein 1 |
| chr13_-_40887444 | 0.04 |
ENSMUST00000224665.2
|
Tfap2a
|
transcription factor AP-2, alpha |
| chrX_-_73472477 | 0.04 |
ENSMUST00000143521.2
|
G6pdx
|
glucose-6-phosphate dehydrogenase X-linked |
| chr16_-_22847829 | 0.04 |
ENSMUST00000100046.9
|
Kng2
|
kininogen 2 |
| chr12_-_113542610 | 0.04 |
ENSMUST00000195468.6
ENSMUST00000103442.3 |
Ighv5-2
|
immunoglobulin heavy variable 5-2 |
| chr7_+_13467422 | 0.04 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr7_+_27147475 | 0.04 |
ENSMUST00000133750.8
|
Blvrb
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr19_-_33602652 | 0.04 |
ENSMUST00000124230.3
|
Gm8978
|
predicted gene 8978 |
| chr10_-_99595498 | 0.04 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr18_-_72484126 | 0.04 |
ENSMUST00000114943.11
|
Dcc
|
deleted in colorectal carcinoma |
| chr7_+_78563150 | 0.04 |
ENSMUST00000118867.8
|
Isg20
|
interferon-stimulated protein |
| chr11_-_84761637 | 0.03 |
ENSMUST00000168434.8
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr18_+_52958382 | 0.03 |
ENSMUST00000238707.2
|
Gm50457
|
predicted gene, 50457 |
| chr15_+_31224460 | 0.03 |
ENSMUST00000044524.16
|
Dap
|
death-associated protein |
| chr11_+_61896161 | 0.03 |
ENSMUST00000201624.4
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr16_-_88759753 | 0.03 |
ENSMUST00000179707.3
|
Krtap16-3
|
keratin associated protein 16-3 |
| chrX_-_103244784 | 0.03 |
ENSMUST00000118314.8
|
Nexmif
|
neurite extension and migration factor |
| chr19_+_34268053 | 0.03 |
ENSMUST00000025691.13
|
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr14_-_44112974 | 0.03 |
ENSMUST00000179200.2
|
Ear1
|
eosinophil-associated, ribonuclease A family, member 1 |
| chrX_+_55939732 | 0.03 |
ENSMUST00000136396.8
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr5_+_90708962 | 0.03 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr17_+_36134398 | 0.03 |
ENSMUST00000173493.8
ENSMUST00000173147.8 |
Flot1
|
flotillin 1 |
| chr11_-_99383938 | 0.03 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr18_+_37818263 | 0.03 |
ENSMUST00000194418.2
|
Pcdhga4
|
protocadherin gamma subfamily A, 4 |
| chr17_-_57535003 | 0.03 |
ENSMUST00000177046.2
ENSMUST00000024988.15 |
C3
|
complement component 3 |
| chr4_-_132459762 | 0.03 |
ENSMUST00000045550.5
|
Xkr8
|
X-linked Kx blood group related 8 |
| chr6_+_129374441 | 0.03 |
ENSMUST00000112081.9
ENSMUST00000112079.3 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr19_-_7779943 | 0.03 |
ENSMUST00000120522.8
|
Slc22a26
|
solute carrier family 22 (organic cation transporter), member 26 |
| chr19_-_57227742 | 0.03 |
ENSMUST00000111559.8
|
Ablim1
|
actin-binding LIM protein 1 |
| chr11_-_89732091 | 0.03 |
ENSMUST00000238273.3
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr17_-_79076487 | 0.03 |
ENSMUST00000233850.2
|
Heatr5b
|
HEAT repeat containing 5B |
| chr2_-_65397809 | 0.03 |
ENSMUST00000066432.12
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr1_+_75456173 | 0.03 |
ENSMUST00000113575.9
ENSMUST00000148980.2 ENSMUST00000050899.7 ENSMUST00000187411.2 |
Tmem198
|
transmembrane protein 198 |
| chr16_-_22847760 | 0.03 |
ENSMUST00000039338.13
|
Kng2
|
kininogen 2 |
| chr2_+_111610658 | 0.03 |
ENSMUST00000054004.2
|
Olfr1302
|
olfactory receptor 1302 |
| chrX_-_103244703 | 0.03 |
ENSMUST00000087879.11
|
Nexmif
|
neurite extension and migration factor |
| chr1_-_37535202 | 0.03 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr2_-_32277773 | 0.03 |
ENSMUST00000050785.14
|
Lcn2
|
lipocalin 2 |
| chr13_+_24798591 | 0.03 |
ENSMUST00000091694.10
|
Ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr18_-_43925932 | 0.03 |
ENSMUST00000237926.2
ENSMUST00000096570.4 |
Gm94
|
predicted gene 94 |
| chr7_+_16550198 | 0.02 |
ENSMUST00000108495.9
|
Strn4
|
striatin, calmodulin binding protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpa_Cebpg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.5 | GO:0071727 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.5 | GO:1904253 | lipid transport involved in lipid storage(GO:0010877) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.2 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.4 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.2 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.1 | 0.2 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.2 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.1 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.2 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0070162 | adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) |
| 0.0 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.0 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:1902988 | regulation of neuronal signal transduction(GO:1902847) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:1990696 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.5 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 0.3 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.3 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0042602 | riboflavin reductase (NADPH) activity(GO:0042602) |
| 0.0 | 0.2 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |