Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
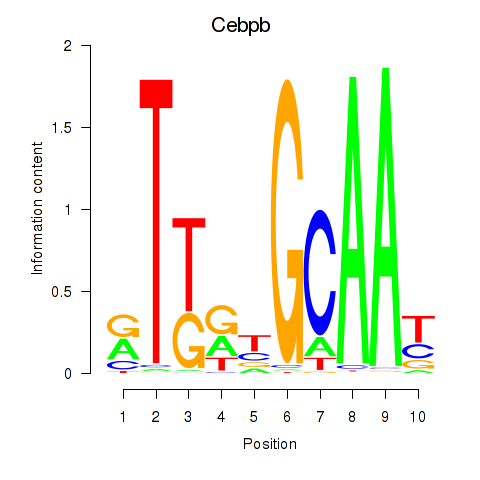
Results for Cebpb
Z-value: 1.73
Transcription factors associated with Cebpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cebpb
|
ENSMUSG00000056501.4 | CCAAT/enhancer binding protein (C/EBP), beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpb | mm39_v1_chr2_+_167530835_167530894 | -0.03 | 9.6e-01 | Click! |
Activity profile of Cebpb motif
Sorted Z-values of Cebpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_109678685 | 3.35 |
ENSMUST00000112022.5
|
Camp
|
cathelicidin antimicrobial peptide |
| chr19_-_11618192 | 2.60 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr19_-_11618165 | 2.57 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr11_-_75313350 | 1.46 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr11_-_75313412 | 1.42 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr9_+_110248815 | 1.17 |
ENSMUST00000035061.9
|
Ngp
|
neutrophilic granule protein |
| chr17_+_48653429 | 1.12 |
ENSMUST00000024791.15
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr17_-_56428968 | 1.09 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr3_-_50398027 | 0.96 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr17_+_48653493 | 0.96 |
ENSMUST00000113237.4
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr9_+_110867807 | 0.95 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr6_-_56878854 | 0.95 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr9_+_107957640 | 0.88 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr9_+_107957621 | 0.88 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr4_-_136626073 | 0.87 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr17_+_87270504 | 0.79 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr4_+_152123772 | 0.78 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr4_+_150322151 | 0.77 |
ENSMUST00000141931.2
|
Eno1
|
enolase 1, alpha non-neuron |
| chr10_+_80765900 | 0.72 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr3_+_106393348 | 0.70 |
ENSMUST00000183271.2
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr1_+_134110142 | 0.69 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr3_+_87814147 | 0.68 |
ENSMUST00000159492.8
|
Hdgf
|
heparin binding growth factor |
| chr7_-_46365108 | 0.65 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr7_-_107696793 | 0.65 |
ENSMUST00000217304.2
|
Olfr482
|
olfactory receptor 482 |
| chr15_-_103218876 | 0.64 |
ENSMUST00000079824.6
|
Gpr84
|
G protein-coupled receptor 84 |
| chr17_+_47908025 | 0.64 |
ENSMUST00000183206.2
|
Ccnd3
|
cyclin D3 |
| chr13_-_53135064 | 0.64 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr7_+_120634834 | 0.62 |
ENSMUST00000207351.2
|
Mettl9
|
methyltransferase like 9 |
| chr14_+_51181956 | 0.61 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr13_+_25127127 | 0.60 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr13_+_49835576 | 0.59 |
ENSMUST00000165316.8
ENSMUST00000047363.14 |
Iars
|
isoleucine-tRNA synthetase |
| chr1_+_40123858 | 0.59 |
ENSMUST00000027243.13
|
Il1r2
|
interleukin 1 receptor, type II |
| chr7_-_45480200 | 0.56 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr9_-_39405284 | 0.55 |
ENSMUST00000077757.7
ENSMUST00000215065.3 ENSMUST00000216316.3 |
Olfr44
|
olfactory receptor 44 |
| chr8_-_46664321 | 0.53 |
ENSMUST00000034049.5
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr5_+_135038006 | 0.53 |
ENSMUST00000111216.8
ENSMUST00000046999.12 |
Abhd11
|
abhydrolase domain containing 11 |
| chr6_+_40605758 | 0.52 |
ENSMUST00000202636.4
ENSMUST00000201148.4 ENSMUST00000071535.10 |
Mgam
|
maltase-glucoamylase |
| chr5_+_115417725 | 0.52 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr2_+_119181703 | 0.49 |
ENSMUST00000028780.4
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr2_-_3513783 | 0.48 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr2_-_32278245 | 0.48 |
ENSMUST00000192241.2
|
Lcn2
|
lipocalin 2 |
| chr10_+_79722081 | 0.47 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr18_-_64649497 | 0.47 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr7_+_30463175 | 0.46 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr6_-_11907392 | 0.45 |
ENSMUST00000204084.3
ENSMUST00000031637.8 ENSMUST00000204978.3 ENSMUST00000204714.2 |
Ndufa4
|
Ndufa4, mitochondrial complex associated |
| chr7_-_45083688 | 0.45 |
ENSMUST00000210439.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr7_-_126873219 | 0.45 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr11_-_45943138 | 0.44 |
ENSMUST00000093169.3
|
Gm12166
|
predicted gene 12166 |
| chr5_+_31212165 | 0.44 |
ENSMUST00000202795.4
ENSMUST00000201182.4 ENSMUST00000200953.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr19_-_15902292 | 0.44 |
ENSMUST00000025542.10
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr8_+_111760521 | 0.43 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr13_+_49835667 | 0.43 |
ENSMUST00000172254.3
|
Iars
|
isoleucine-tRNA synthetase |
| chr3_+_96011810 | 0.42 |
ENSMUST00000132980.8
ENSMUST00000138206.8 ENSMUST00000090785.9 ENSMUST00000035519.12 |
Otud7b
|
OTU domain containing 7B |
| chr5_+_31212110 | 0.41 |
ENSMUST00000013773.12
ENSMUST00000201838.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr15_+_41694317 | 0.41 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr17_+_34131696 | 0.41 |
ENSMUST00000174146.3
|
Daxx
|
Fas death domain-associated protein |
| chr8_+_86219191 | 0.40 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr4_+_63262775 | 0.39 |
ENSMUST00000030044.3
|
Orm1
|
orosomucoid 1 |
| chr11_-_3321307 | 0.39 |
ENSMUST00000101640.10
ENSMUST00000101642.10 |
Limk2
|
LIM motif-containing protein kinase 2 |
| chr11_+_49141339 | 0.36 |
ENSMUST00000101293.11
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr8_+_26298502 | 0.35 |
ENSMUST00000033979.6
|
Star
|
steroidogenic acute regulatory protein |
| chr2_-_164198427 | 0.35 |
ENSMUST00000109367.10
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr1_+_171173252 | 0.35 |
ENSMUST00000006579.5
|
Pfdn2
|
prefoldin 2 |
| chr1_+_171173237 | 0.35 |
ENSMUST00000135941.8
|
Pfdn2
|
prefoldin 2 |
| chr3_+_109481223 | 0.34 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr17_+_87270707 | 0.34 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr18_-_64649620 | 0.33 |
ENSMUST00000025483.11
ENSMUST00000237400.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr4_-_108690741 | 0.33 |
ENSMUST00000102740.8
ENSMUST00000102741.8 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr3_+_20011201 | 0.33 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr11_-_120521382 | 0.32 |
ENSMUST00000106181.8
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr1_-_131204651 | 0.32 |
ENSMUST00000161764.8
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr1_-_78465479 | 0.32 |
ENSMUST00000190441.2
ENSMUST00000170217.8 ENSMUST00000188247.7 ENSMUST00000068333.14 |
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr10_+_127919142 | 0.31 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr19_+_55240357 | 0.31 |
ENSMUST00000225551.2
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr10_+_61556371 | 0.31 |
ENSMUST00000080099.6
|
Aifm2
|
apoptosis-inducing factor, mitochondrion-associated 2 |
| chr17_+_28910302 | 0.30 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr2_-_152185901 | 0.30 |
ENSMUST00000040312.7
|
Trib3
|
tribbles pseudokinase 3 |
| chr3_-_121608859 | 0.30 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr9_+_5298669 | 0.29 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr3_+_31150982 | 0.29 |
ENSMUST00000118204.2
|
Skil
|
SKI-like |
| chr5_-_129856267 | 0.28 |
ENSMUST00000201394.4
|
Psph
|
phosphoserine phosphatase |
| chr19_-_8690330 | 0.28 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr11_-_87716849 | 0.28 |
ENSMUST00000103177.10
|
Lpo
|
lactoperoxidase |
| chr1_+_55170476 | 0.28 |
ENSMUST00000162553.2
|
Mob4
|
MOB family member 4, phocein |
| chr5_-_137531413 | 0.28 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr4_+_126915104 | 0.28 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr6_-_3968365 | 0.28 |
ENSMUST00000031674.11
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr11_-_120534469 | 0.28 |
ENSMUST00000141254.8
ENSMUST00000170556.8 ENSMUST00000151876.8 ENSMUST00000026133.15 ENSMUST00000139706.2 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr11_+_106107752 | 0.27 |
ENSMUST00000021046.6
|
Ddx42
|
DEAD box helicase 42 |
| chr7_-_44181477 | 0.27 |
ENSMUST00000098483.9
ENSMUST00000035323.6 |
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr14_-_54648057 | 0.27 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr14_+_22069709 | 0.27 |
ENSMUST00000075639.11
|
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr13_-_49462694 | 0.27 |
ENSMUST00000110087.9
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr4_-_133694543 | 0.27 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_-_43558386 | 0.26 |
ENSMUST00000130353.2
|
Tln1
|
talin 1 |
| chr4_-_151129020 | 0.26 |
ENSMUST00000103204.11
|
Per3
|
period circadian clock 3 |
| chr11_-_101442663 | 0.25 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr11_+_49141410 | 0.25 |
ENSMUST00000129588.2
|
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr4_-_130068902 | 0.25 |
ENSMUST00000105998.8
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr3_-_121608809 | 0.25 |
ENSMUST00000197383.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr17_+_27276262 | 0.24 |
ENSMUST00000049308.9
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr11_-_72441054 | 0.24 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chr5_+_93416091 | 0.24 |
ENSMUST00000121127.2
|
Ccng2
|
cyclin G2 |
| chr5_-_123126550 | 0.24 |
ENSMUST00000086200.11
ENSMUST00000156474.8 |
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr10_-_88192852 | 0.23 |
ENSMUST00000020249.2
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr4_-_107928567 | 0.23 |
ENSMUST00000106701.2
|
Scp2
|
sterol carrier protein 2, liver |
| chr11_-_86977576 | 0.22 |
ENSMUST00000020801.14
|
Smg8
|
smg-8 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr11_-_120520954 | 0.22 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr9_-_103107460 | 0.22 |
ENSMUST00000165296.8
ENSMUST00000112645.8 |
Trf
|
transferrin |
| chr13_+_95460110 | 0.22 |
ENSMUST00000221807.2
|
Zbed3
|
zinc finger, BED type containing 3 |
| chr11_-_84761472 | 0.21 |
ENSMUST00000018547.9
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr2_+_25313240 | 0.21 |
ENSMUST00000134259.8
ENSMUST00000100320.5 |
Fut7
|
fucosyltransferase 7 |
| chr7_+_109617456 | 0.21 |
ENSMUST00000084731.5
|
Ipo7
|
importin 7 |
| chr2_+_103858168 | 0.20 |
ENSMUST00000111135.8
ENSMUST00000111136.8 ENSMUST00000102565.4 |
Fbxo3
|
F-box protein 3 |
| chr6_-_87786736 | 0.20 |
ENSMUST00000032134.9
|
Rab43
|
RAB43, member RAS oncogene family |
| chr7_-_109092834 | 0.19 |
ENSMUST00000106739.8
|
Trim66
|
tripartite motif-containing 66 |
| chr19_-_8796288 | 0.19 |
ENSMUST00000153281.2
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr14_-_55108384 | 0.19 |
ENSMUST00000146642.2
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr11_-_102120953 | 0.19 |
ENSMUST00000107150.8
ENSMUST00000156337.2 ENSMUST00000107151.9 ENSMUST00000107152.9 |
Hdac5
|
histone deacetylase 5 |
| chr2_+_103858066 | 0.19 |
ENSMUST00000028603.10
|
Fbxo3
|
F-box protein 3 |
| chr13_-_117161921 | 0.18 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr13_+_51254852 | 0.18 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr11_-_102120917 | 0.18 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr7_+_45084300 | 0.18 |
ENSMUST00000211150.2
|
Gys1
|
glycogen synthase 1, muscle |
| chr6_-_123266820 | 0.18 |
ENSMUST00000032239.11
ENSMUST00000177367.8 |
Clec4e
|
C-type lectin domain family 4, member e |
| chr14_+_31887739 | 0.17 |
ENSMUST00000111994.10
ENSMUST00000168114.8 ENSMUST00000168034.8 |
Ncoa4
|
nuclear receptor coactivator 4 |
| chr8_+_94763826 | 0.17 |
ENSMUST00000109556.9
ENSMUST00000093301.9 ENSMUST00000060632.8 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr14_-_54647647 | 0.16 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr19_+_12438125 | 0.16 |
ENSMUST00000081035.9
|
Mpeg1
|
macrophage expressed gene 1 |
| chr2_-_129213050 | 0.16 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr17_+_29333116 | 0.16 |
ENSMUST00000233717.2
ENSMUST00000141239.2 |
Rab44
|
RAB44, member RAS oncogene family |
| chr7_+_18915086 | 0.16 |
ENSMUST00000120595.8
ENSMUST00000048502.10 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr11_-_6150411 | 0.15 |
ENSMUST00000066496.10
|
Nudcd3
|
NudC domain containing 3 |
| chr15_+_37233280 | 0.15 |
ENSMUST00000161405.8
ENSMUST00000022895.15 ENSMUST00000161532.2 |
Grhl2
|
grainyhead like transcription factor 2 |
| chr7_-_45084012 | 0.15 |
ENSMUST00000107771.12
ENSMUST00000211666.2 |
Ruvbl2
|
RuvB-like protein 2 |
| chr11_-_84761538 | 0.15 |
ENSMUST00000170741.2
ENSMUST00000172405.8 ENSMUST00000100686.10 ENSMUST00000108081.9 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr11_-_84761637 | 0.15 |
ENSMUST00000168434.8
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr2_+_15531281 | 0.15 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr1_+_55170411 | 0.14 |
ENSMUST00000027122.14
|
Mob4
|
MOB family member 4, phocein |
| chr1_-_171173168 | 0.14 |
ENSMUST00000156856.8
ENSMUST00000111296.8 |
Nit1
|
nitrilase 1 |
| chr12_-_113843161 | 0.14 |
ENSMUST00000103451.5
|
Ighv2-9
|
immunoglobulin heavy variable 2-9 |
| chr1_-_79838897 | 0.14 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr9_-_71678814 | 0.13 |
ENSMUST00000122065.2
ENSMUST00000121322.8 ENSMUST00000072899.9 |
Cgnl1
|
cingulin-like 1 |
| chr17_-_57501170 | 0.13 |
ENSMUST00000005976.8
|
Tnfsf14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr3_+_146110709 | 0.13 |
ENSMUST00000129978.2
|
Ssx2ip
|
synovial sarcoma, X 2 interacting protein |
| chr19_-_8796677 | 0.13 |
ENSMUST00000096751.11
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr4_+_155646807 | 0.13 |
ENSMUST00000030939.14
|
Nadk
|
NAD kinase |
| chrX_-_56384089 | 0.12 |
ENSMUST00000033468.11
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr13_-_117162041 | 0.12 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chrX_-_133501874 | 0.12 |
ENSMUST00000033621.8
|
Gla
|
galactosidase, alpha |
| chr3_+_66127330 | 0.12 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr19_+_6451667 | 0.12 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr10_-_99595498 | 0.12 |
ENSMUST00000056085.6
|
Csl
|
citrate synthase like |
| chr10_-_8632519 | 0.12 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr1_+_55170390 | 0.11 |
ENSMUST00000159311.8
ENSMUST00000162364.8 |
Mob4
|
MOB family member 4, phocein |
| chr7_+_30475819 | 0.11 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr14_-_68893253 | 0.11 |
ENSMUST00000225767.3
ENSMUST00000111072.8 ENSMUST00000022642.6 ENSMUST00000224039.2 |
Adam28
|
a disintegrin and metallopeptidase domain 28 |
| chr17_+_35863025 | 0.11 |
ENSMUST00000044804.8
|
Cdsn
|
corneodesmosin |
| chr11_-_99383938 | 0.11 |
ENSMUST00000006969.8
|
Krt23
|
keratin 23 |
| chr8_+_10056654 | 0.10 |
ENSMUST00000033892.9
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr7_+_78563150 | 0.10 |
ENSMUST00000118867.8
|
Isg20
|
interferon-stimulated protein |
| chr4_-_148172423 | 0.10 |
ENSMUST00000030865.9
|
Agtrap
|
angiotensin II, type I receptor-associated protein |
| chr10_+_127126643 | 0.10 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr12_-_101784727 | 0.10 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr12_+_24622274 | 0.09 |
ENSMUST00000085553.13
|
Grhl1
|
grainyhead like transcription factor 1 |
| chr4_-_130253703 | 0.09 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr14_-_49304110 | 0.09 |
ENSMUST00000162175.9
|
Exoc5
|
exocyst complex component 5 |
| chr7_+_18915136 | 0.09 |
ENSMUST00000144054.8
ENSMUST00000141718.8 |
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr1_+_128079543 | 0.08 |
ENSMUST00000189317.3
|
R3hdm1
|
R3H domain containing 1 |
| chr3_+_103739877 | 0.08 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr13_+_83723255 | 0.08 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr12_+_76884182 | 0.08 |
ENSMUST00000041008.10
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr11_+_110956980 | 0.08 |
ENSMUST00000042970.3
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr12_-_70160491 | 0.08 |
ENSMUST00000222237.2
|
Nin
|
ninein |
| chr2_+_172235820 | 0.07 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr6_-_116050081 | 0.07 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr7_-_45109075 | 0.07 |
ENSMUST00000210864.2
|
Ftl1
|
ferritin light polypeptide 1 |
| chr3_+_20011251 | 0.07 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr8_+_11547506 | 0.07 |
ENSMUST00000177955.8
ENSMUST00000033901.11 ENSMUST00000178721.2 |
Naxd
|
NAD(P)HX dehydratase |
| chr3_+_20011405 | 0.06 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr12_-_101785307 | 0.06 |
ENSMUST00000021603.9
|
Fbln5
|
fibulin 5 |
| chr9_-_103107495 | 0.06 |
ENSMUST00000035158.16
|
Trf
|
transferrin |
| chr9_+_121211820 | 0.06 |
ENSMUST00000209995.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr15_-_76191301 | 0.06 |
ENSMUST00000171340.9
ENSMUST00000023222.13 ENSMUST00000164189.2 |
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr13_+_16186410 | 0.06 |
ENSMUST00000042603.14
|
Inhba
|
inhibin beta-A |
| chr15_+_59520199 | 0.06 |
ENSMUST00000067543.8
|
Trib1
|
tribbles pseudokinase 1 |
| chr4_+_124635635 | 0.06 |
ENSMUST00000094782.10
ENSMUST00000153837.8 ENSMUST00000154229.2 |
Inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr7_+_45084257 | 0.06 |
ENSMUST00000003964.17
|
Gys1
|
glycogen synthase 1, muscle |
| chr1_-_131204422 | 0.06 |
ENSMUST00000159195.2
|
Ikbke
|
inhibitor of kappaB kinase epsilon |
| chr4_-_123458465 | 0.06 |
ENSMUST00000238731.2
|
Macf1
|
microtubule-actin crosslinking factor 1 |
| chr16_-_21814190 | 0.06 |
ENSMUST00000231766.2
ENSMUST00000074230.12 |
Liph
|
lipase, member H |
| chr1_-_164281344 | 0.06 |
ENSMUST00000193367.2
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr5_+_75312939 | 0.06 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr14_-_55950545 | 0.05 |
ENSMUST00000002389.9
ENSMUST00000226907.2 |
Tgm1
|
transglutaminase 1, K polypeptide |
| chr10_-_6930376 | 0.05 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr18_+_44237474 | 0.05 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chrX_+_94942639 | 0.05 |
ENSMUST00000082183.8
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr14_+_22069877 | 0.05 |
ENSMUST00000161249.8
ENSMUST00000159777.8 ENSMUST00000162540.2 |
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr2_+_119863909 | 0.05 |
ENSMUST00000126150.2
|
Pla2g4b
|
phospholipase A2, group IVB (cytosolic) |
| chr18_+_4994600 | 0.04 |
ENSMUST00000140448.8
|
Svil
|
supervillin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.4 | 4.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.3 | 1.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 | 1.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 0.8 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 2.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 | 0.6 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 1.8 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.2 | 0.8 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.2 | 0.5 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.2 | 0.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 0.6 | GO:0006507 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.6 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.5 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.8 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.6 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.4 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.1 | 1.0 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.4 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 0.3 | GO:0050705 | regulation of interleukin-1 alpha secretion(GO:0050705) |
| 0.1 | 0.4 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.3 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.2 | GO:1901373 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.1 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 0.2 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.2 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.3 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.3 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.3 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 1.0 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.2 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.4 | GO:0090051 | regulation of skeletal muscle fiber development(GO:0048742) negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.0 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 4.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.3 | 0.8 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 0.8 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.2 | 2.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.2 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.4 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.6 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.8 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.5 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.3 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.7 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.2 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.3 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.1 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.9 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 4.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |