Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
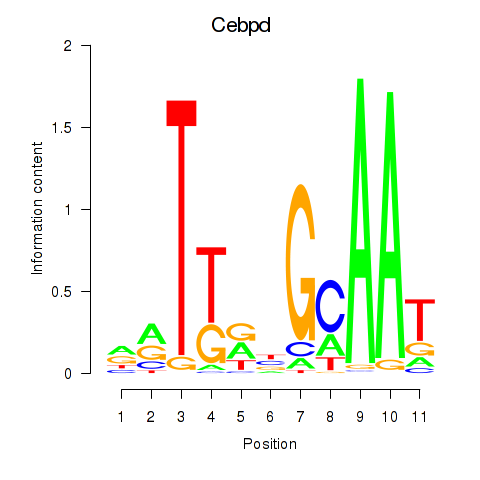
Results for Cebpd
Z-value: 0.93
Transcription factors associated with Cebpd
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cebpd
|
ENSMUSG00000071637.6 | CCAAT/enhancer binding protein (C/EBP), delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpd | mm39_v1_chr16_+_15705141_15705156 | -0.98 | 4.2e-03 | Click! |
Activity profile of Cebpd motif
Sorted Z-values of Cebpd motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_75313412 | 0.96 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr11_-_75313350 | 0.93 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr14_+_67953547 | 0.68 |
ENSMUST00000078053.13
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr17_-_56428968 | 0.66 |
ENSMUST00000041357.9
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr4_-_136626073 | 0.57 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr17_+_48653429 | 0.52 |
ENSMUST00000024791.15
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr17_+_87270504 | 0.49 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr4_+_152123772 | 0.44 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr17_+_48653493 | 0.43 |
ENSMUST00000113237.4
|
Trem2
|
triggering receptor expressed on myeloid cells 2 |
| chr17_-_36343573 | 0.42 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr17_+_37581103 | 0.39 |
ENSMUST00000038580.7
|
H2-M3
|
histocompatibility 2, M region locus 3 |
| chr7_-_126873219 | 0.38 |
ENSMUST00000082428.6
|
Sephs2
|
selenophosphate synthetase 2 |
| chr8_-_46664321 | 0.36 |
ENSMUST00000034049.5
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr7_-_46365108 | 0.32 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr5_+_115417725 | 0.30 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr7_-_45480200 | 0.29 |
ENSMUST00000107723.9
ENSMUST00000131384.3 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr5_-_100867520 | 0.29 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr4_+_63262775 | 0.28 |
ENSMUST00000030044.3
|
Orm1
|
orosomucoid 1 |
| chr4_-_108690741 | 0.27 |
ENSMUST00000102740.8
ENSMUST00000102741.8 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr11_+_106107752 | 0.26 |
ENSMUST00000021046.6
|
Ddx42
|
DEAD box helicase 42 |
| chr2_-_3513783 | 0.26 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr11_-_106107132 | 0.24 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr6_-_11907392 | 0.23 |
ENSMUST00000204084.3
ENSMUST00000031637.8 ENSMUST00000204978.3 ENSMUST00000204714.2 |
Ndufa4
|
Ndufa4, mitochondrial complex associated |
| chr16_-_65359406 | 0.23 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr5_-_129856267 | 0.21 |
ENSMUST00000201394.4
|
Psph
|
phosphoserine phosphatase |
| chr11_-_72441054 | 0.21 |
ENSMUST00000021154.7
|
Spns3
|
spinster homolog 3 |
| chrX_+_158086253 | 0.20 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr17_+_87270707 | 0.20 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr19_+_55240357 | 0.19 |
ENSMUST00000225551.2
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr7_+_30463175 | 0.17 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr14_+_22069709 | 0.17 |
ENSMUST00000075639.11
|
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr6_-_3968365 | 0.15 |
ENSMUST00000031674.11
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr14_+_67953584 | 0.15 |
ENSMUST00000145542.8
ENSMUST00000125212.2 |
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr9_-_103107460 | 0.15 |
ENSMUST00000165296.8
ENSMUST00000112645.8 |
Trf
|
transferrin |
| chr3_+_20011201 | 0.14 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr4_-_43558386 | 0.14 |
ENSMUST00000130353.2
|
Tln1
|
talin 1 |
| chr2_-_129213050 | 0.13 |
ENSMUST00000028881.14
|
Il1b
|
interleukin 1 beta |
| chr11_-_86977576 | 0.11 |
ENSMUST00000020801.14
|
Smg8
|
smg-8 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr5_-_147831610 | 0.11 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr7_-_44181477 | 0.10 |
ENSMUST00000098483.9
ENSMUST00000035323.6 |
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related) |
| chr2_+_172235820 | 0.09 |
ENSMUST00000109136.3
ENSMUST00000228775.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chrX_-_133501874 | 0.09 |
ENSMUST00000033621.8
|
Gla
|
galactosidase, alpha |
| chr2_+_15531281 | 0.09 |
ENSMUST00000146205.3
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr13_-_117161921 | 0.09 |
ENSMUST00000223949.2
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr3_+_66127330 | 0.07 |
ENSMUST00000029421.6
|
Ptx3
|
pentraxin related gene |
| chr6_-_87786736 | 0.07 |
ENSMUST00000032134.9
|
Rab43
|
RAB43, member RAS oncogene family |
| chr8_+_94763826 | 0.07 |
ENSMUST00000109556.9
ENSMUST00000093301.9 ENSMUST00000060632.8 |
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr4_-_130253703 | 0.07 |
ENSMUST00000134159.3
|
Zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr13_-_117162041 | 0.06 |
ENSMUST00000022239.8
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr19_-_8796288 | 0.06 |
ENSMUST00000153281.2
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr10_-_6930376 | 0.06 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr5_-_129856237 | 0.05 |
ENSMUST00000118268.9
|
Psph
|
phosphoserine phosphatase |
| chr18_+_44237474 | 0.04 |
ENSMUST00000081271.7
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr7_-_45109075 | 0.04 |
ENSMUST00000210864.2
|
Ftl1
|
ferritin light polypeptide 1 |
| chr9_-_103107495 | 0.04 |
ENSMUST00000035158.16
|
Trf
|
transferrin |
| chr1_-_164281344 | 0.04 |
ENSMUST00000193367.2
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr3_+_20011405 | 0.04 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chrX_+_133501928 | 0.04 |
ENSMUST00000074950.11
ENSMUST00000113203.2 ENSMUST00000113202.8 ENSMUST00000050331.13 ENSMUST00000059297.6 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chr3_+_20011251 | 0.04 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr8_+_10056654 | 0.03 |
ENSMUST00000033892.9
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr18_+_44237577 | 0.03 |
ENSMUST00000239465.2
|
Spink12
|
serine peptidase inhibitor, Kazal type 12 |
| chr19_-_8796677 | 0.03 |
ENSMUST00000096751.11
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr19_-_50667079 | 0.03 |
ENSMUST00000209413.2
ENSMUST00000072685.13 ENSMUST00000164039.9 |
Sorcs1
|
sortilin-related VPS10 domain containing receptor 1 |
| chr5_+_114284585 | 0.03 |
ENSMUST00000102582.8
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr18_-_52662728 | 0.02 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
| chr2_-_88559941 | 0.02 |
ENSMUST00000099815.2
|
Olfr1197
|
olfactory receptor 1197 |
| chr18_-_52662917 | 0.02 |
ENSMUST00000171470.8
|
Lox
|
lysyl oxidase |
| chr2_-_88844406 | 0.02 |
ENSMUST00000099800.2
|
Olfr1216
|
olfactory receptor 1216 |
| chr2_-_110781268 | 0.02 |
ENSMUST00000099623.10
|
Ano3
|
anoctamin 3 |
| chr7_+_26006594 | 0.02 |
ENSMUST00000098657.5
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr14_-_68771138 | 0.01 |
ENSMUST00000022640.8
|
Adam7
|
a disintegrin and metallopeptidase domain 7 |
| chr7_+_13467422 | 0.01 |
ENSMUST00000086148.8
|
Sult2a2
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 |
| chr5_+_8010445 | 0.01 |
ENSMUST00000115421.3
|
Steap4
|
STEAP family member 4 |
| chr6_-_116050081 | 0.01 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr14_-_45715308 | 0.01 |
ENSMUST00000141424.2
|
Fermt2
|
fermitin family member 2 |
| chr16_-_22847808 | 0.01 |
ENSMUST00000115349.9
|
Kng2
|
kininogen 2 |
| chr1_-_106980033 | 0.01 |
ENSMUST00000112717.3
|
Serpinb3a
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A |
| chr14_+_22069877 | 0.01 |
ENSMUST00000161249.8
ENSMUST00000159777.8 ENSMUST00000162540.2 |
Lrmda
|
leucine rich melanocyte differentiation associated |
| chr18_-_43925932 | 0.01 |
ENSMUST00000237926.2
ENSMUST00000096570.4 |
Gm94
|
predicted gene 94 |
| chr8_+_10056631 | 0.01 |
ENSMUST00000207792.3
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr1_-_107088844 | 0.01 |
ENSMUST00000086694.11
|
Serpinb3b
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3B |
| chr15_-_101759212 | 0.00 |
ENSMUST00000023790.5
|
Krt1
|
keratin 1 |
| chr6_+_17463748 | 0.00 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr1_+_164624200 | 0.00 |
ENSMUST00000027861.6
|
Dpt
|
dermatopontin |
| chr17_-_36679309 | 0.00 |
ENSMUST00000073546.5
|
H2-M10.3
|
histocompatibility 2, M region locus 10.3 |
| chr2_+_14828903 | 0.00 |
ENSMUST00000193800.6
|
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr2_-_25360043 | 0.00 |
ENSMUST00000114251.8
|
Ptgds
|
prostaglandin D2 synthase (brain) |
| chr9_-_48516447 | 0.00 |
ENSMUST00000034808.12
ENSMUST00000119426.2 |
Nnmt
|
nicotinamide N-methyltransferase |
| chr2_-_25359752 | 0.00 |
ENSMUST00000114259.3
ENSMUST00000015234.13 |
Ptgds
|
prostaglandin D2 synthase (brain) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cebpd
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0002588 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.3 | 0.8 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.2 | 1.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0070447 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 | 0.7 | GO:1904376 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.7 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) regulation of mitotic spindle assembly(GO:1901673) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.8 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.4 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0031544 | peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |