Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Clock
Z-value: 0.32
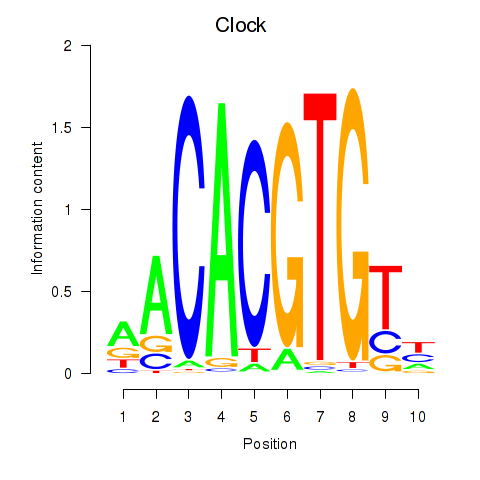
Transcription factors associated with Clock
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Clock
|
ENSMUSG00000029238.12 | circadian locomotor output cycles kaput |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Clock | mm39_v1_chr5_-_76452577_76452644 | 0.75 | 1.5e-01 | Click! |
Activity profile of Clock motif
Sorted Z-values of Clock motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_68986043 | 0.33 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr7_-_68398989 | 0.33 |
ENSMUST00000048068.15
|
Arrdc4
|
arrestin domain containing 4 |
| chr13_-_12355604 | 0.23 |
ENSMUST00000168193.8
ENSMUST00000064204.14 |
Actn2
|
actinin alpha 2 |
| chr4_-_57143437 | 0.20 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr10_+_22034460 | 0.19 |
ENSMUST00000181645.8
ENSMUST00000105522.9 |
Raet1e
H60b
|
retinoic acid early transcript 1E histocompatibility 60b |
| chr12_+_24701273 | 0.18 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr8_-_46452896 | 0.17 |
ENSMUST00000053558.10
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr7_-_68398917 | 0.16 |
ENSMUST00000118110.3
|
Arrdc4
|
arrestin domain containing 4 |
| chr4_+_107825529 | 0.11 |
ENSMUST00000106713.5
ENSMUST00000238795.2 |
Slc1a7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr4_-_133484080 | 0.11 |
ENSMUST00000008024.7
|
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr8_+_108669276 | 0.10 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3 |
| chr15_-_102533870 | 0.09 |
ENSMUST00000184485.8
ENSMUST00000185070.8 ENSMUST00000184616.8 ENSMUST00000108828.9 |
Atf7
|
activating transcription factor 7 |
| chr17_+_47922497 | 0.09 |
ENSMUST00000024778.3
|
Med20
|
mediator complex subunit 20 |
| chr2_-_130748259 | 0.09 |
ENSMUST00000128176.2
ENSMUST00000133766.2 ENSMUST00000135110.2 ENSMUST00000146975.2 |
A730017L22Rik
4930402H24Rik
|
RIKEN cDNA A730017L22 gene RIKEN cDNA 4930402H24 gene |
| chr13_+_20369377 | 0.09 |
ENSMUST00000221067.2
|
Elmo1
|
engulfment and cell motility 1 |
| chr2_-_38816229 | 0.08 |
ENSMUST00000076275.11
ENSMUST00000142130.2 |
Nr6a1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr19_-_29782907 | 0.08 |
ENSMUST00000175726.8
|
9930021J03Rik
|
RIKEN cDNA 9930021J03 gene |
| chr7_-_144493560 | 0.08 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr9_-_57513510 | 0.07 |
ENSMUST00000215487.2
ENSMUST00000045068.10 |
Cplx3
|
complexin 3 |
| chr6_-_91450486 | 0.07 |
ENSMUST00000040835.9
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr5_-_74692327 | 0.07 |
ENSMUST00000072857.13
ENSMUST00000113542.9 ENSMUST00000151474.3 |
Scfd2
|
Sec1 family domain containing 2 |
| chr16_+_43710163 | 0.07 |
ENSMUST00000132859.8
ENSMUST00000178400.9 |
Ccdc191
|
coiled-coil domain containing 191 |
| chrX_+_7439839 | 0.07 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr5_-_46014809 | 0.07 |
ENSMUST00000190036.7
ENSMUST00000189859.7 ENSMUST00000186633.3 ENSMUST00000016026.14 ENSMUST00000045586.13 ENSMUST00000238522.2 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr16_+_3854806 | 0.06 |
ENSMUST00000137748.8
ENSMUST00000006136.11 ENSMUST00000157044.8 ENSMUST00000120009.8 |
Dnase1
|
deoxyribonuclease I |
| chr4_+_44756553 | 0.06 |
ENSMUST00000107824.9
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr19_+_46044972 | 0.06 |
ENSMUST00000111899.8
ENSMUST00000099392.10 ENSMUST00000062322.11 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr17_+_35220252 | 0.06 |
ENSMUST00000174260.8
|
Vars
|
valyl-tRNA synthetase |
| chr16_+_35590745 | 0.06 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr17_-_66384017 | 0.06 |
ENSMUST00000150766.2
ENSMUST00000038116.13 |
Ankrd12
|
ankyrin repeat domain 12 |
| chr2_+_48839505 | 0.05 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chrX_+_8137881 | 0.05 |
ENSMUST00000115590.2
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr17_-_28705055 | 0.05 |
ENSMUST00000233870.2
|
Fkbp5
|
FK506 binding protein 5 |
| chr11_+_74721733 | 0.05 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein |
| chr19_+_9995557 | 0.05 |
ENSMUST00000113161.10
ENSMUST00000238672.2 ENSMUST00000117641.8 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr7_-_29204812 | 0.05 |
ENSMUST00000183096.8
ENSMUST00000085809.11 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr2_+_32285908 | 0.05 |
ENSMUST00000028162.5
|
Ptges2
|
prostaglandin E synthase 2 |
| chr4_+_44756608 | 0.05 |
ENSMUST00000143385.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr6_+_17463819 | 0.05 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr2_-_130266162 | 0.05 |
ENSMUST00000089581.11
|
Pced1a
|
PC-esterase domain containing 1A |
| chr7_-_126294902 | 0.05 |
ENSMUST00000144897.2
|
Slx1b
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr11_-_118180895 | 0.05 |
ENSMUST00000144153.8
|
Usp36
|
ubiquitin specific peptidase 36 |
| chr14_+_4230658 | 0.05 |
ENSMUST00000225491.2
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr11_-_118181044 | 0.05 |
ENSMUST00000106296.9
ENSMUST00000092382.10 |
Usp36
|
ubiquitin specific peptidase 36 |
| chr7_-_46445085 | 0.05 |
ENSMUST00000123725.2
|
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr19_+_46045675 | 0.05 |
ENSMUST00000126127.8
ENSMUST00000147640.2 |
Pprc1
|
peroxisome proliferative activated receptor, gamma, coactivator-related 1 |
| chr15_+_80556023 | 0.04 |
ENSMUST00000023044.7
|
Fam83f
|
family with sequence similarity 83, member F |
| chr12_-_32000534 | 0.04 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1 |
| chr15_+_103148824 | 0.04 |
ENSMUST00000036004.16
ENSMUST00000087351.9 ENSMUST00000231141.2 |
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr11_+_106680062 | 0.04 |
ENSMUST00000103068.10
ENSMUST00000018516.11 |
Cep95
|
centrosomal protein 95 |
| chr3_-_101195213 | 0.04 |
ENSMUST00000029456.5
|
Cd2
|
CD2 antigen |
| chr4_-_155947819 | 0.04 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr2_+_14234198 | 0.04 |
ENSMUST00000028045.4
|
Mrc1
|
mannose receptor, C type 1 |
| chr18_-_60981981 | 0.04 |
ENSMUST00000177172.8
ENSMUST00000175934.8 ENSMUST00000176630.8 |
Tcof1
|
treacle ribosome biogenesis factor 1 |
| chr14_-_78970160 | 0.04 |
ENSMUST00000226342.3
|
Dgkh
|
diacylglycerol kinase, eta |
| chr7_+_66489465 | 0.04 |
ENSMUST00000098382.10
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr9_+_14695933 | 0.04 |
ENSMUST00000034405.11
ENSMUST00000115632.10 ENSMUST00000147305.2 |
Mre11a
|
MRE11A homolog A, double strand break repair nuclease |
| chr16_-_44978986 | 0.03 |
ENSMUST00000180636.8
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr19_-_29790352 | 0.03 |
ENSMUST00000099525.5
|
Ranbp6
|
RAN binding protein 6 |
| chr5_-_137784912 | 0.03 |
ENSMUST00000031740.16
|
Mepce
|
methylphosphate capping enzyme |
| chr9_+_40712562 | 0.03 |
ENSMUST00000117557.8
|
Hspa8
|
heat shock protein 8 |
| chr3_-_108117754 | 0.03 |
ENSMUST00000117784.8
ENSMUST00000119650.8 ENSMUST00000117409.8 |
Atxn7l2
|
ataxin 7-like 2 |
| chr2_+_18059982 | 0.03 |
ENSMUST00000028076.15
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr1_-_166237341 | 0.03 |
ENSMUST00000135673.8
ENSMUST00000169324.8 ENSMUST00000128861.3 |
Pogk
|
pogo transposable element with KRAB domain |
| chr1_+_181180183 | 0.03 |
ENSMUST00000161880.8
ENSMUST00000027795.14 |
Cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr4_+_32657105 | 0.03 |
ENSMUST00000071642.11
ENSMUST00000178134.2 |
Mdn1
|
midasin AAA ATPase 1 |
| chr8_+_91555449 | 0.03 |
ENSMUST00000109614.9
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr5_+_146168020 | 0.03 |
ENSMUST00000161181.8
ENSMUST00000161652.8 ENSMUST00000031640.15 ENSMUST00000159467.2 |
Cdk8
|
cyclin-dependent kinase 8 |
| chr11_+_69231589 | 0.03 |
ENSMUST00000218008.2
ENSMUST00000151617.3 |
Rnf227
|
ring finger protein 227 |
| chr10_+_69048914 | 0.03 |
ENSMUST00000163497.8
ENSMUST00000164212.8 ENSMUST00000067908.14 |
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr7_+_66489500 | 0.03 |
ENSMUST00000107478.9
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr18_+_42644552 | 0.03 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr4_+_97665992 | 0.03 |
ENSMUST00000107062.9
ENSMUST00000052018.12 ENSMUST00000107057.8 |
Nfia
|
nuclear factor I/A |
| chr7_+_19311212 | 0.03 |
ENSMUST00000108453.2
|
Zfp296
|
zinc finger protein 296 |
| chr5_+_3391477 | 0.02 |
ENSMUST00000199156.2
|
Cdk6
|
cyclin-dependent kinase 6 |
| chr7_-_46445305 | 0.02 |
ENSMUST00000107653.8
ENSMUST00000107654.8 ENSMUST00000014562.14 ENSMUST00000152759.8 |
Hps5
|
HPS5, biogenesis of lysosomal organelles complex 2 subunit 2 |
| chr6_+_17463925 | 0.02 |
ENSMUST00000115442.8
|
Met
|
met proto-oncogene |
| chr4_-_131802606 | 0.02 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr16_-_43709968 | 0.02 |
ENSMUST00000023387.14
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr15_+_81695615 | 0.02 |
ENSMUST00000023024.8
|
Tef
|
thyrotroph embryonic factor |
| chr2_-_32584132 | 0.02 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr5_+_45650821 | 0.02 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr3_-_58599812 | 0.02 |
ENSMUST00000070368.8
|
Siah2
|
siah E3 ubiquitin protein ligase 2 |
| chr16_+_43710299 | 0.02 |
ENSMUST00000151183.2
|
Ccdc191
|
coiled-coil domain containing 191 |
| chr4_-_131802561 | 0.02 |
ENSMUST00000105970.8
ENSMUST00000105975.8 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr12_-_71183371 | 0.02 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr2_-_118380149 | 0.02 |
ENSMUST00000090219.13
|
Bmf
|
BCL2 modifying factor |
| chr2_+_115412148 | 0.02 |
ENSMUST00000166472.8
ENSMUST00000110918.3 |
Cdin1
|
CDAN1 interacting nuclease 1 |
| chr8_+_121854566 | 0.02 |
ENSMUST00000181609.2
|
Foxl1
|
forkhead box L1 |
| chr12_+_17594795 | 0.02 |
ENSMUST00000171737.3
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr8_-_37081091 | 0.02 |
ENSMUST00000033923.14
|
Dlc1
|
deleted in liver cancer 1 |
| chr16_-_44979013 | 0.02 |
ENSMUST00000023344.10
|
Slc35a5
|
solute carrier family 35, member A5 |
| chr1_-_13061333 | 0.02 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr17_-_47922374 | 0.02 |
ENSMUST00000024783.9
|
Bysl
|
bystin-like |
| chrX_-_94488394 | 0.02 |
ENSMUST00000084535.6
|
Amer1
|
APC membrane recruitment 1 |
| chr12_+_102094977 | 0.02 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr10_+_69048464 | 0.02 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr11_-_51647204 | 0.02 |
ENSMUST00000109092.8
ENSMUST00000064297.5 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr19_+_26600820 | 0.02 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_+_70735751 | 0.02 |
ENSMUST00000177731.8
ENSMUST00000108533.10 ENSMUST00000081362.13 ENSMUST00000178245.2 |
Rabep1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr2_-_168048984 | 0.02 |
ENSMUST00000088001.6
|
Adnp
|
activity-dependent neuroprotective protein |
| chr9_+_43978290 | 0.01 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr5_+_135197228 | 0.01 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr5_+_135197137 | 0.01 |
ENSMUST00000031692.12
|
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr2_-_163261439 | 0.01 |
ENSMUST00000046908.10
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chrX_-_47297436 | 0.01 |
ENSMUST00000037960.11
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr11_+_74540284 | 0.01 |
ENSMUST00000117818.2
ENSMUST00000092915.12 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr4_-_145041710 | 0.01 |
ENSMUST00000030339.13
|
Tnfrsf8
|
tumor necrosis factor receptor superfamily, member 8 |
| chr17_-_24428351 | 0.01 |
ENSMUST00000024931.6
|
Ntn3
|
netrin 3 |
| chr11_+_72332167 | 0.01 |
ENSMUST00000045633.6
|
Mybbp1a
|
MYB binding protein (P160) 1a |
| chr11_-_101676076 | 0.01 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr5_+_77099154 | 0.01 |
ENSMUST00000031160.16
ENSMUST00000120912.8 ENSMUST00000117536.8 |
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chr16_+_18695787 | 0.01 |
ENSMUST00000120532.9
ENSMUST00000004222.14 |
Hira
|
histone cell cycle regulator |
| chr7_+_127345909 | 0.01 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr6_+_17463748 | 0.01 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr14_-_20718337 | 0.01 |
ENSMUST00000057090.12
ENSMUST00000117386.2 |
Synpo2l
|
synaptopodin 2-like |
| chr2_-_48839218 | 0.01 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr12_+_71183622 | 0.01 |
ENSMUST00000149564.8
|
2700049A03Rik
|
RIKEN cDNA 2700049A03 gene |
| chr18_+_35686424 | 0.01 |
ENSMUST00000235679.2
ENSMUST00000235176.2 ENSMUST00000235801.2 ENSMUST00000237592.2 ENSMUST00000237230.2 ENSMUST00000237589.2 |
Snhg4
Snhg4
|
small nucleolar RNA host gene 4 small nucleolar RNA host gene 4 |
| chr6_-_77956499 | 0.01 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chrX_+_8137620 | 0.01 |
ENSMUST00000033512.11
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr18_+_67597929 | 0.01 |
ENSMUST00000025411.9
|
Prelid3a
|
PRELI domain containing 3A |
| chr13_-_76091931 | 0.01 |
ENSMUST00000022078.12
ENSMUST00000109606.3 |
Rhobtb3
|
Rho-related BTB domain containing 3 |
| chr12_+_59178072 | 0.01 |
ENSMUST00000176464.8
ENSMUST00000170992.9 ENSMUST00000176322.8 |
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr9_-_14695801 | 0.01 |
ENSMUST00000214979.2
ENSMUST00000216037.2 ENSMUST00000214456.2 |
Ankrd49
|
ankyrin repeat domain 49 |
| chrX_+_8137372 | 0.01 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr13_-_63712140 | 0.01 |
ENSMUST00000195756.6
|
Ptch1
|
patched 1 |
| chr11_+_78069528 | 0.01 |
ENSMUST00000108338.2
|
Tlcd1
|
TLC domain containing 1 |
| chr6_-_99703344 | 0.01 |
ENSMUST00000008273.8
ENSMUST00000101120.11 ENSMUST00000203738.2 |
Prok2
|
prokineticin 2 |
| chr3_+_41519289 | 0.01 |
ENSMUST00000168086.7
|
Jade1
|
jade family PHD finger 1 |
| chr11_-_51647290 | 0.01 |
ENSMUST00000109097.9
|
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr11_-_100650566 | 0.01 |
ENSMUST00000107361.9
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr6_+_125073108 | 0.01 |
ENSMUST00000112390.8
|
Chd4
|
chromodomain helicase DNA binding protein 4 |
| chr1_+_59724108 | 0.01 |
ENSMUST00000027174.10
ENSMUST00000190231.7 ENSMUST00000191142.7 ENSMUST00000185772.7 |
Nop58
|
NOP58 ribonucleoprotein |
| chr7_-_98010478 | 0.01 |
ENSMUST00000094161.11
ENSMUST00000164726.8 ENSMUST00000206414.2 ENSMUST00000167405.3 |
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr1_-_181039509 | 0.01 |
ENSMUST00000162819.9
ENSMUST00000237749.2 |
Wdr26
|
WD repeat domain 26 |
| chr19_-_41969558 | 0.01 |
ENSMUST00000026168.9
ENSMUST00000171561.8 |
Mms19
|
MMS19 cytosolic iron-sulfur assembly component |
| chr8_+_123912976 | 0.01 |
ENSMUST00000019422.6
|
Dpep1
|
dipeptidase 1 |
| chr17_+_43327412 | 0.00 |
ENSMUST00000024708.6
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr11_-_100650768 | 0.00 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr5_+_77099229 | 0.00 |
ENSMUST00000141687.2
|
Paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase |
| chrX_-_92675719 | 0.00 |
ENSMUST00000006856.3
|
Pola1
|
polymerase (DNA directed), alpha 1 |
| chr11_-_60307847 | 0.00 |
ENSMUST00000108721.9
ENSMUST00000239016.2 |
Atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr7_-_34914675 | 0.00 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr12_-_32000209 | 0.00 |
ENSMUST00000176084.2
ENSMUST00000176103.8 ENSMUST00000167458.9 |
Hbp1
|
high mobility group box transcription factor 1 |
| chrX_+_5959507 | 0.00 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr5_-_77099406 | 0.00 |
ENSMUST00000140076.2
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr10_+_116137277 | 0.00 |
ENSMUST00000092167.7
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr7_+_25358589 | 0.00 |
ENSMUST00000079634.8
ENSMUST00000206561.2 |
Exosc5
|
exosome component 5 |
| chr3_+_135531409 | 0.00 |
ENSMUST00000180196.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr3_-_95789338 | 0.00 |
ENSMUST00000161994.2
|
Ciart
|
circadian associated repressor of transcription |
| chr3_-_87702469 | 0.00 |
ENSMUST00000029712.5
|
Ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr3_+_40663285 | 0.00 |
ENSMUST00000091184.9
|
Slc25a31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chrX_-_36127891 | 0.00 |
ENSMUST00000115249.10
ENSMUST00000115248.10 |
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr11_-_77380492 | 0.00 |
ENSMUST00000037593.14
ENSMUST00000092892.10 |
Ankrd13b
|
ankyrin repeat domain 13b |
| chr1_-_186438177 | 0.00 |
ENSMUST00000045288.14
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr11_-_106107132 | 0.00 |
ENSMUST00000002043.10
|
Ccdc47
|
coiled-coil domain containing 47 |
| chr14_-_20231871 | 0.00 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr4_+_106954794 | 0.00 |
ENSMUST00000221740.2
|
Cdcp2
|
CUB domain containing protein 2 |
| chr11_+_76108434 | 0.00 |
ENSMUST00000040806.5
|
Dbil5
|
diazepam binding inhibitor-like 5 |
| chr11_-_76108316 | 0.00 |
ENSMUST00000102500.5
|
Gemin4
|
gem nuclear organelle associated protein 4 |
| chr3_+_145281941 | 0.00 |
ENSMUST00000199033.5
ENSMUST00000098534.9 ENSMUST00000200574.5 ENSMUST00000196413.5 ENSMUST00000197604.3 |
Znhit6
|
zinc finger, HIT type 6 |
| chr5_+_120254530 | 0.00 |
ENSMUST00000031590.12
|
Rbm19
|
RNA binding motif protein 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Clock
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.1 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0002654 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.0 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.0 | GO:0097212 | protein targeting to vacuole involved in autophagy(GO:0071211) lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |