Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
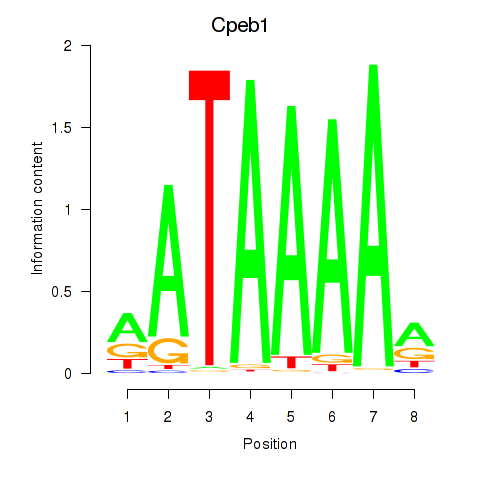
Results for Cpeb1
Z-value: 1.22
Transcription factors associated with Cpeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cpeb1
|
ENSMUSG00000025586.18 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cpeb1 | mm39_v1_chr7_-_81104423_81104512 | 0.46 | 4.4e-01 | Click! |
Activity profile of Cpeb1 motif
Sorted Z-values of Cpeb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_18053158 | 2.68 |
ENSMUST00000066885.6
|
Skida1
|
SKI/DACH domain containing 1 |
| chr13_-_23806530 | 2.42 |
ENSMUST00000062045.4
|
H1f4
|
H1.4 linker histone, cluster member |
| chr13_-_23882437 | 2.02 |
ENSMUST00000102967.3
|
H4c3
|
H4 clustered histone 3 |
| chr13_+_22220000 | 2.01 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr6_+_79794899 | 1.84 |
ENSMUST00000179797.3
|
Gm20594
|
predicted gene, 20594 |
| chr13_-_23945189 | 1.78 |
ENSMUST00000102964.4
|
H4c1
|
H4 clustered histone 1 |
| chr3_+_96152813 | 1.52 |
ENSMUST00000078756.7
ENSMUST00000090779.4 |
H2ac18
Gm20634
|
H2A clustered histone 18 predicted gene 20634 |
| chr6_-_136781406 | 1.51 |
ENSMUST00000179285.3
|
H4f16
|
H4 histone 16 |
| chr2_-_18053595 | 1.47 |
ENSMUST00000142856.2
|
Skida1
|
SKI/DACH domain containing 1 |
| chr3_-_96147592 | 1.37 |
ENSMUST00000074976.8
|
H2ac19
|
H2A clustered histone 19 |
| chr3_+_96177010 | 1.09 |
ENSMUST00000051089.4
ENSMUST00000177113.2 |
Gm42743
H2bc18
|
predicted gene 42743 H2B clustered histone 18 |
| chr17_+_36227869 | 0.81 |
ENSMUST00000151664.8
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr2_-_165997551 | 0.81 |
ENSMUST00000109249.9
ENSMUST00000146497.9 |
Sulf2
|
sulfatase 2 |
| chr13_-_22219738 | 0.78 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chrM_+_2743 | 0.77 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr4_-_117354249 | 0.68 |
ENSMUST00000030439.15
|
Rnf220
|
ring finger protein 220 |
| chr6_-_52190299 | 0.62 |
ENSMUST00000128102.4
|
Gm28308
|
predicted gene 28308 |
| chr5_+_90907207 | 0.59 |
ENSMUST00000031318.6
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5 |
| chr3_-_58792633 | 0.59 |
ENSMUST00000055636.13
ENSMUST00000072551.7 ENSMUST00000051408.8 |
Clrn1
|
clarin 1 |
| chr13_+_23868175 | 0.59 |
ENSMUST00000018246.6
|
H2bc4
|
H2B clustered histone 4 |
| chr4_-_44168252 | 0.56 |
ENSMUST00000145760.8
ENSMUST00000128426.8 |
Rnf38
|
ring finger protein 38 |
| chr17_-_36208265 | 0.53 |
ENSMUST00000148721.8
|
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr5_-_142803135 | 0.51 |
ENSMUST00000198181.2
|
Tnrc18
|
trinucleotide repeat containing 18 |
| chr2_+_84818538 | 0.51 |
ENSMUST00000028466.12
|
Prg3
|
proteoglycan 3 |
| chr16_-_4831349 | 0.51 |
ENSMUST00000201077.2
ENSMUST00000202281.4 ENSMUST00000090453.9 ENSMUST00000023191.17 |
Rogdi
|
rogdi homolog |
| chr18_+_61178211 | 0.50 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr19_+_8828132 | 0.48 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr8_+_85449632 | 0.47 |
ENSMUST00000098571.5
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr16_-_48592372 | 0.47 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1 |
| chr1_-_63153414 | 0.45 |
ENSMUST00000153992.2
ENSMUST00000165066.8 ENSMUST00000172416.8 ENSMUST00000137511.8 |
Ino80d
|
INO80 complex subunit D |
| chr6_-_99005835 | 0.45 |
ENSMUST00000154163.9
|
Foxp1
|
forkhead box P1 |
| chr2_-_51039112 | 0.44 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr5_-_116427003 | 0.44 |
ENSMUST00000086483.4
ENSMUST00000050178.13 |
Ccdc60
|
coiled-coil domain containing 60 |
| chr16_+_36097313 | 0.44 |
ENSMUST00000232150.2
|
Stfa1
|
stefin A1 |
| chr11_+_69011230 | 0.43 |
ENSMUST00000024543.3
|
Hes7
|
hes family bHLH transcription factor 7 |
| chrM_+_14138 | 0.43 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b |
| chr13_-_23867924 | 0.43 |
ENSMUST00000171127.4
|
H2ac6
|
H2A clustered histone 6 |
| chrM_-_14061 | 0.43 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chrX_+_158623460 | 0.42 |
ENSMUST00000112451.8
ENSMUST00000112453.9 |
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr19_+_4203603 | 0.41 |
ENSMUST00000236632.2
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C polypeptide-associated protein |
| chr2_+_87404246 | 0.41 |
ENSMUST00000213315.2
ENSMUST00000214773.2 |
Olfr1129
|
olfactory receptor 1129 |
| chr16_-_23807602 | 0.39 |
ENSMUST00000023151.6
|
Bcl6
|
B cell leukemia/lymphoma 6 |
| chr1_-_171023798 | 0.39 |
ENSMUST00000111332.2
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr6_-_66759611 | 0.38 |
ENSMUST00000226457.2
|
Vmn1r38
|
vomeronasal 1 receptor 38 |
| chr2_-_165997179 | 0.38 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr2_-_102731691 | 0.38 |
ENSMUST00000111192.3
ENSMUST00000111190.9 ENSMUST00000111198.9 ENSMUST00000111191.9 ENSMUST00000060516.14 ENSMUST00000099673.9 ENSMUST00000005218.15 ENSMUST00000111194.8 |
Cd44
|
CD44 antigen |
| chr10_-_68114543 | 0.37 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr16_-_55659194 | 0.37 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr8_-_5155347 | 0.37 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr1_+_131936022 | 0.37 |
ENSMUST00000146432.2
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr11_-_118246332 | 0.36 |
ENSMUST00000017610.10
|
Timp2
|
tissue inhibitor of metalloproteinase 2 |
| chr10_+_34265969 | 0.36 |
ENSMUST00000105511.2
|
Col10a1
|
collagen, type X, alpha 1 |
| chr8_-_64659004 | 0.36 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr6_-_85310393 | 0.35 |
ENSMUST00000059034.13
ENSMUST00000045846.12 ENSMUST00000113788.2 |
Sfxn5
|
sideroflexin 5 |
| chr11_-_110142565 | 0.35 |
ENSMUST00000044003.14
|
Abca6
|
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr5_-_66308421 | 0.34 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr11_-_57983936 | 0.34 |
ENSMUST00000218429.2
ENSMUST00000219877.2 |
Gm12248
|
predicted gene 12248 |
| chr7_+_79460475 | 0.33 |
ENSMUST00000107394.3
|
Mesp2
|
mesoderm posterior 2 |
| chr18_+_69726431 | 0.33 |
ENSMUST00000201091.4
ENSMUST00000201037.4 ENSMUST00000114977.5 |
Tcf4
|
transcription factor 4 |
| chr12_-_99359265 | 0.33 |
ENSMUST00000177451.8
|
Foxn3
|
forkhead box N3 |
| chr5_-_84565218 | 0.32 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr1_+_17672117 | 0.32 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chrX_-_156198282 | 0.31 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr12_+_40495951 | 0.30 |
ENSMUST00000037488.8
|
Dock4
|
dedicator of cytokinesis 4 |
| chr17_-_45047521 | 0.30 |
ENSMUST00000113572.9
|
Runx2
|
runt related transcription factor 2 |
| chr19_+_44191704 | 0.30 |
ENSMUST00000026220.7
|
Scd3
|
stearoyl-coenzyme A desaturase 3 |
| chr12_+_84616536 | 0.29 |
ENSMUST00000021665.12
ENSMUST00000169934.4 |
Vsx2
|
visual system homeobox 2 |
| chr7_-_103734672 | 0.29 |
ENSMUST00000057104.7
|
Olfr645
|
olfactory receptor 645 |
| chr18_+_69726654 | 0.29 |
ENSMUST00000200921.4
|
Tcf4
|
transcription factor 4 |
| chrX_+_41239872 | 0.29 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2 |
| chr2_+_152578164 | 0.28 |
ENSMUST00000038368.9
ENSMUST00000109824.2 |
Id1
|
inhibitor of DNA binding 1, HLH protein |
| chr16_+_65317389 | 0.27 |
ENSMUST00000176330.8
ENSMUST00000004964.15 ENSMUST00000176038.8 |
Pou1f1
|
POU domain, class 1, transcription factor 1 |
| chr2_+_92015780 | 0.27 |
ENSMUST00000128781.9
ENSMUST00000111291.9 |
Phf21a
|
PHD finger protein 21A |
| chr11_+_29413734 | 0.27 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr2_-_160208977 | 0.26 |
ENSMUST00000099126.5
|
Mafb
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
| chr17_-_34847354 | 0.26 |
ENSMUST00000167097.9
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr11_-_103158190 | 0.25 |
ENSMUST00000021324.3
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14 |
| chr9_-_63929308 | 0.25 |
ENSMUST00000179458.2
ENSMUST00000041029.6 |
Smad6
|
SMAD family member 6 |
| chr13_+_81031512 | 0.25 |
ENSMUST00000099356.10
|
Arrdc3
|
arrestin domain containing 3 |
| chr15_+_76238632 | 0.25 |
ENSMUST00000208833.3
|
Gm35339
|
predicted gene, 35339 |
| chr17_+_36227433 | 0.24 |
ENSMUST00000087211.9
|
Ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr13_+_83720484 | 0.24 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_45003270 | 0.24 |
ENSMUST00000202935.4
ENSMUST00000068415.11 ENSMUST00000127520.8 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_-_82045800 | 0.24 |
ENSMUST00000235015.2
ENSMUST00000163123.3 |
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr11_-_4897991 | 0.24 |
ENSMUST00000093369.5
|
Nefh
|
neurofilament, heavy polypeptide |
| chr2_+_48839505 | 0.23 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr13_+_41040657 | 0.23 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr5_-_66308666 | 0.23 |
ENSMUST00000201561.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr1_-_128520002 | 0.23 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr11_-_70545450 | 0.23 |
ENSMUST00000018437.3
|
Pfn1
|
profilin 1 |
| chr5_+_149335214 | 0.23 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr7_-_30251680 | 0.23 |
ENSMUST00000215288.2
ENSMUST00000108165.8 ENSMUST00000153594.3 |
Proser3
|
proline and serine rich 3 |
| chr3_-_146388165 | 0.23 |
ENSMUST00000124931.8
ENSMUST00000147113.2 |
Samd13
|
sterile alpha motif domain containing 13 |
| chr10_+_127256736 | 0.22 |
ENSMUST00000064793.13
|
R3hdm2
|
R3H domain containing 2 |
| chrX_-_151110425 | 0.22 |
ENSMUST00000195280.3
|
Kantr
|
Kdm5c adjacent non-coding transcript |
| chrM_+_5319 | 0.21 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I |
| chr2_-_45002902 | 0.21 |
ENSMUST00000076836.13
ENSMUST00000176732.8 ENSMUST00000200844.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr10_+_29087602 | 0.21 |
ENSMUST00000092627.6
|
9330159F19Rik
|
RIKEN cDNA 9330159F19 gene |
| chr12_-_25146078 | 0.20 |
ENSMUST00000222667.2
ENSMUST00000020974.7 |
Id2
|
inhibitor of DNA binding 2 |
| chr13_+_37529184 | 0.20 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr18_+_75151780 | 0.20 |
ENSMUST00000236220.2
ENSMUST00000039608.9 ENSMUST00000235692.2 |
Dym
|
dymeclin |
| chr12_-_44257109 | 0.20 |
ENSMUST00000015049.5
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr7_-_28079678 | 0.20 |
ENSMUST00000051241.7
|
Zfp36
|
zinc finger protein 36 |
| chr1_-_84817976 | 0.20 |
ENSMUST00000190067.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr10_+_19232281 | 0.20 |
ENSMUST00000053225.7
|
Olig3
|
oligodendrocyte transcription factor 3 |
| chr3_+_84500854 | 0.20 |
ENSMUST00000062623.4
|
Tigd4
|
tigger transposable element derived 4 |
| chr12_-_44256843 | 0.19 |
ENSMUST00000220421.2
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chrX_+_102400061 | 0.19 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr9_+_44684248 | 0.19 |
ENSMUST00000128150.8
|
Ift46
|
intraflagellar transport 46 |
| chr2_-_87838612 | 0.19 |
ENSMUST00000215457.2
|
Olfr1160
|
olfactory receptor 1160 |
| chr2_+_164782642 | 0.19 |
ENSMUST00000137626.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr12_+_38830081 | 0.19 |
ENSMUST00000095767.11
|
Etv1
|
ets variant 1 |
| chr3_+_105821450 | 0.19 |
ENSMUST00000198080.5
ENSMUST00000199977.2 |
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr4_+_62537750 | 0.19 |
ENSMUST00000084521.11
ENSMUST00000107424.2 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr12_+_110413677 | 0.19 |
ENSMUST00000220509.2
|
Ppp2r5c
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr8_-_29709652 | 0.19 |
ENSMUST00000168630.4
|
Unc5d
|
unc-5 netrin receptor D |
| chr17_+_18108086 | 0.18 |
ENSMUST00000149944.2
|
Fpr2
|
formyl peptide receptor 2 |
| chr11_+_75422516 | 0.18 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr9_+_56982622 | 0.18 |
ENSMUST00000167715.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast) |
| chr9_+_44684285 | 0.17 |
ENSMUST00000239014.2
|
Ift46
|
intraflagellar transport 46 |
| chr2_+_111984716 | 0.17 |
ENSMUST00000214063.2
ENSMUST00000217533.2 |
Olfr1318
|
olfactory receptor 1318 |
| chr16_+_57369595 | 0.17 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr10_-_30076488 | 0.17 |
ENSMUST00000216853.2
|
Cenpw
|
centromere protein W |
| chrX_-_48823936 | 0.17 |
ENSMUST00000215373.3
|
Olfr1321
|
olfactory receptor 1321 |
| chr3_+_60503051 | 0.17 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr9_-_116004265 | 0.17 |
ENSMUST00000061101.12
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr7_-_16549394 | 0.17 |
ENSMUST00000206259.2
|
Fkrp
|
fukutin related protein |
| chr11_+_78006391 | 0.17 |
ENSMUST00000155571.2
|
Fam222b
|
family with sequence similarity 222, member B |
| chr17_-_10501816 | 0.17 |
ENSMUST00000233684.2
|
Qk
|
quaking, KH domain containing RNA binding |
| chr4_+_150366028 | 0.17 |
ENSMUST00000105682.9
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr14_-_67106037 | 0.16 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr11_+_67477347 | 0.16 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr16_+_44913974 | 0.16 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr15_-_50753792 | 0.16 |
ENSMUST00000185183.2
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chrX_+_109857866 | 0.16 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr8_+_34582184 | 0.16 |
ENSMUST00000095345.5
|
Mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr1_+_178356678 | 0.15 |
ENSMUST00000161017.8
|
Kif26b
|
kinesin family member 26B |
| chr15_+_61857226 | 0.15 |
ENSMUST00000161976.8
ENSMUST00000022971.8 |
Myc
|
myelocytomatosis oncogene |
| chr9_-_40366966 | 0.15 |
ENSMUST00000165104.8
ENSMUST00000045682.7 |
Gramd1b
|
GRAM domain containing 1B |
| chr19_+_13890894 | 0.15 |
ENSMUST00000216623.2
ENSMUST00000216835.2 |
Olfr1505
|
olfactory receptor 1505 |
| chr2_+_14179324 | 0.15 |
ENSMUST00000077517.9
|
Tmem236
|
transmembrane protein 236 |
| chr14_+_28740162 | 0.15 |
ENSMUST00000055662.4
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr6_-_52203146 | 0.15 |
ENSMUST00000114425.3
|
Hoxa9
|
homeobox A9 |
| chr19_+_60878802 | 0.15 |
ENSMUST00000236876.2
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chrX_+_74460234 | 0.15 |
ENSMUST00000033544.14
|
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr18_-_22983794 | 0.14 |
ENSMUST00000092015.11
ENSMUST00000069215.13 |
Nol4
|
nucleolar protein 4 |
| chr11_-_78313043 | 0.14 |
ENSMUST00000001122.6
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr12_-_31763859 | 0.14 |
ENSMUST00000057783.6
ENSMUST00000236002.2 ENSMUST00000174480.3 ENSMUST00000176710.2 |
Gpr22
|
G protein-coupled receptor 22 |
| chr7_+_4122523 | 0.14 |
ENSMUST00000119661.8
ENSMUST00000129423.8 |
Ttyh1
|
tweety family member 1 |
| chr13_+_83720457 | 0.14 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr9_-_85209340 | 0.14 |
ENSMUST00000187711.2
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr11_+_31822211 | 0.14 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_-_53159885 | 0.14 |
ENSMUST00000030010.4
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr17_-_46638915 | 0.14 |
ENSMUST00000167360.8
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr2_-_148285450 | 0.14 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr19_-_20931566 | 0.14 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr17_+_36179273 | 0.13 |
ENSMUST00000190496.2
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr11_+_115921129 | 0.13 |
ENSMUST00000021116.12
ENSMUST00000106452.2 |
Unk
|
unkempt family zinc finger |
| chr4_-_136563154 | 0.13 |
ENSMUST00000105846.9
ENSMUST00000059287.14 ENSMUST00000105845.9 |
Ephb2
|
Eph receptor B2 |
| chr9_+_121780054 | 0.13 |
ENSMUST00000043011.9
ENSMUST00000214536.3 ENSMUST00000215990.3 |
Gask1a
|
golgi associated kinase 1A |
| chr15_-_36609208 | 0.13 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_+_142052569 | 0.13 |
ENSMUST00000078497.15
ENSMUST00000105953.10 ENSMUST00000179658.8 ENSMUST00000105954.10 ENSMUST00000105952.10 ENSMUST00000105955.8 ENSMUST00000074187.13 ENSMUST00000169299.9 ENSMUST00000105957.10 ENSMUST00000180152.8 ENSMUST00000105950.11 ENSMUST00000105958.10 ENSMUST00000105949.8 |
Tnnt3
|
troponin T3, skeletal, fast |
| chr11_+_108811168 | 0.13 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr18_-_6490808 | 0.13 |
ENSMUST00000028100.13
ENSMUST00000050542.6 |
Epc1
|
enhancer of polycomb homolog 1 |
| chr1_-_22031718 | 0.13 |
ENSMUST00000029667.13
ENSMUST00000173058.8 ENSMUST00000173404.2 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr8_+_88978600 | 0.13 |
ENSMUST00000154115.2
|
Tent4b
|
terminal nucleotidyltransferase 4B |
| chr5_+_122534900 | 0.13 |
ENSMUST00000196969.5
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr7_-_97387429 | 0.12 |
ENSMUST00000206389.2
|
Aqp11
|
aquaporin 11 |
| chr7_+_128290204 | 0.12 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr7_-_92319096 | 0.12 |
ENSMUST00000208255.2
ENSMUST00000208058.2 |
Pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr7_-_109215754 | 0.12 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr11_-_106679671 | 0.12 |
ENSMUST00000123339.2
|
Ddx5
|
DEAD box helicase 5 |
| chr2_+_89830489 | 0.12 |
ENSMUST00000111508.9
ENSMUST00000131072.3 |
Olfr1262
|
olfactory receptor 1262 |
| chr3_+_5815863 | 0.12 |
ENSMUST00000192045.2
|
Gm8797
|
predicted pseudogene 8797 |
| chr1_+_153750081 | 0.12 |
ENSMUST00000055314.4
|
Teddm1b
|
transmembrane epididymal protein 1B |
| chr9_-_85209162 | 0.12 |
ENSMUST00000034802.15
|
Tent5a
|
terminal nucleotidyltransferase 5A |
| chr1_-_138775317 | 0.12 |
ENSMUST00000093486.10
ENSMUST00000046870.13 |
Lhx9
|
LIM homeobox protein 9 |
| chr8_+_106620174 | 0.12 |
ENSMUST00000060167.6
ENSMUST00000118920.2 |
Nrn1l
|
neuritin 1-like |
| chr6_+_149226891 | 0.12 |
ENSMUST00000189837.2
|
Resf1
|
retroelement silencing factor 1 |
| chr11_-_83540175 | 0.12 |
ENSMUST00000001008.6
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr9_-_32454157 | 0.11 |
ENSMUST00000183767.2
|
Fli1
|
Friend leukemia integration 1 |
| chr1_-_69726384 | 0.11 |
ENSMUST00000187184.7
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr14_-_104081827 | 0.11 |
ENSMUST00000022718.11
|
Ednrb
|
endothelin receptor type B |
| chr12_-_57244096 | 0.11 |
ENSMUST00000044634.12
|
Slc25a21
|
solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 |
| chr9_-_44714263 | 0.11 |
ENSMUST00000044694.8
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr7_-_101486983 | 0.11 |
ENSMUST00000185929.2
ENSMUST00000165052.8 |
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr7_+_79675727 | 0.11 |
ENSMUST00000049680.10
|
Zfp710
|
zinc finger protein 710 |
| chr14_-_52516695 | 0.11 |
ENSMUST00000167116.8
ENSMUST00000100631.11 |
Rab2b
|
RAB2B, member RAS oncogene family |
| chr11_+_75422925 | 0.11 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr9_+_54606832 | 0.11 |
ENSMUST00000070070.8
|
Dnaja4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr17_-_46638874 | 0.11 |
ENSMUST00000047970.14
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr4_-_44168339 | 0.11 |
ENSMUST00000045793.15
|
Rnf38
|
ring finger protein 38 |
| chr1_+_128069677 | 0.11 |
ENSMUST00000187023.7
|
R3hdm1
|
R3H domain containing 1 |
| chr9_+_44684206 | 0.11 |
ENSMUST00000002099.11
|
Ift46
|
intraflagellar transport 46 |
| chr6_+_136495818 | 0.11 |
ENSMUST00000186577.7
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr13_+_83672708 | 0.11 |
ENSMUST00000199105.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr10_-_30531768 | 0.10 |
ENSMUST00000092610.12
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr15_+_61857390 | 0.10 |
ENSMUST00000159327.2
ENSMUST00000167731.8 |
Myc
|
myelocytomatosis oncogene |
| chr8_-_34237752 | 0.10 |
ENSMUST00000179364.3
|
Smim18
|
small integral membrane protein 18 |
| chr2_-_75768752 | 0.10 |
ENSMUST00000099996.5
|
Ttc30b
|
tetratricopeptide repeat domain 30B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cpeb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.2 | 1.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 0.5 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.5 | GO:0045575 | basophil activation(GO:0045575) |
| 0.1 | 0.4 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.1 | 0.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of tolerance induction to self antigen(GO:0002649) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 0.3 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.5 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 0.3 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.3 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.2 | GO:1904582 | positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.3 | GO:0090096 | lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.3 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.3 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.6 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.0 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0071313 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.4 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.8 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0055099 | regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.6 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0061181 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0014063 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0030210 | negative regulation of cytokine secretion involved in immune response(GO:0002740) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.0 | 0.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.0 | GO:1903465 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.2 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.4 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.2 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 1.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.0 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.0 | 0.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |