Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
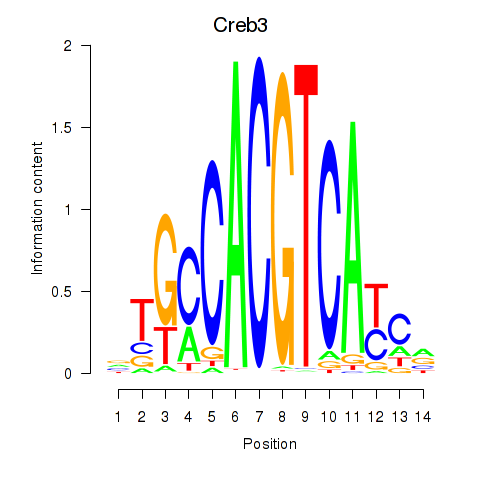
Results for Creb3
Z-value: 1.30
Transcription factors associated with Creb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb3
|
ENSMUSG00000028466.16 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3 | mm39_v1_chr4_+_43562706_43562947 | 0.10 | 8.7e-01 | Click! |
Activity profile of Creb3 motif
Sorted Z-values of Creb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_35658131 | 1.21 |
ENSMUST00000071951.14
ENSMUST00000116598.10 ENSMUST00000078205.14 ENSMUST00000076256.8 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr2_+_80145805 | 0.91 |
ENSMUST00000028392.8
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr6_+_88061464 | 0.70 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr4_-_133972890 | 0.66 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr5_+_45677571 | 0.65 |
ENSMUST00000156481.8
ENSMUST00000119579.3 ENSMUST00000118833.3 |
Med28
|
mediator complex subunit 28 |
| chr11_-_69872050 | 0.59 |
ENSMUST00000108594.8
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr19_-_6886898 | 0.57 |
ENSMUST00000238095.2
|
Prdx5
|
peroxiredoxin 5 |
| chr2_-_105229653 | 0.56 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr7_-_45116197 | 0.56 |
ENSMUST00000211195.2
ENSMUST00000210019.2 |
Bax
|
BCL2-associated X protein |
| chr5_+_52898910 | 0.52 |
ENSMUST00000031081.11
ENSMUST00000031082.8 |
Pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr4_+_47474652 | 0.51 |
ENSMUST00000065678.6
|
Sec61b
|
Sec61 beta subunit |
| chr7_-_45116216 | 0.51 |
ENSMUST00000210392.2
ENSMUST00000211365.2 |
Bax
|
BCL2-associated X protein |
| chr4_+_47474715 | 0.50 |
ENSMUST00000137461.8
ENSMUST00000125622.2 |
Sec61b
|
Sec61 beta subunit |
| chr19_+_8944369 | 0.50 |
ENSMUST00000052248.8
|
Eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr5_-_124490296 | 0.48 |
ENSMUST00000111472.6
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr11_+_52123016 | 0.46 |
ENSMUST00000109072.2
|
Skp1
|
S-phase kinase-associated protein 1 |
| chr11_+_52122836 | 0.45 |
ENSMUST00000037324.12
ENSMUST00000166537.8 |
Skp1
|
S-phase kinase-associated protein 1 |
| chr8_-_106738514 | 0.44 |
ENSMUST00000058579.7
|
Ddx28
|
DEAD box helicase 28 |
| chr7_-_79765042 | 0.42 |
ENSMUST00000206714.2
ENSMUST00000107384.10 |
Idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr16_+_49519561 | 0.42 |
ENSMUST00000046777.11
ENSMUST00000142682.9 |
Ift57
|
intraflagellar transport 57 |
| chr8_+_106738105 | 0.42 |
ENSMUST00000034375.11
|
Dus2
|
dihydrouridine synthase 2 |
| chrX_-_135642025 | 0.41 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr5_+_28276353 | 0.41 |
ENSMUST00000059155.11
|
Insig1
|
insulin induced gene 1 |
| chr9_+_44290832 | 0.41 |
ENSMUST00000161318.8
ENSMUST00000217019.2 ENSMUST00000160902.8 |
Hyou1
|
hypoxia up-regulated 1 |
| chr17_-_56490887 | 0.40 |
ENSMUST00000019723.8
|
Mydgf
|
myeloid derived growth factor |
| chr10_+_128158413 | 0.39 |
ENSMUST00000219836.2
|
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr3_+_89153704 | 0.39 |
ENSMUST00000168900.3
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr11_-_103588605 | 0.38 |
ENSMUST00000021329.14
|
Gosr2
|
golgi SNAP receptor complex member 2 |
| chr18_+_35347983 | 0.38 |
ENSMUST00000235449.2
ENSMUST00000235269.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr5_-_137531463 | 0.38 |
ENSMUST00000170293.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr11_-_120440519 | 0.37 |
ENSMUST00000034913.5
|
Mcrip1
|
MAPK regulated corepressor interacting protein 1 |
| chr10_+_81012465 | 0.37 |
ENSMUST00000047864.11
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr7_-_113853894 | 0.37 |
ENSMUST00000033012.9
|
Copb1
|
coatomer protein complex, subunit beta 1 |
| chr1_-_93373145 | 0.36 |
ENSMUST00000186787.7
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr7_+_138792890 | 0.36 |
ENSMUST00000016124.15
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr13_-_38178059 | 0.35 |
ENSMUST00000225319.2
ENSMUST00000225246.2 ENSMUST00000021864.8 |
Ssr1
|
signal sequence receptor, alpha |
| chr1_+_157334298 | 0.35 |
ENSMUST00000086130.9
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chrX_-_135641869 | 0.35 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr1_+_157334347 | 0.35 |
ENSMUST00000027881.15
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr15_+_6416229 | 0.34 |
ENSMUST00000110664.9
ENSMUST00000110663.9 ENSMUST00000161812.8 ENSMUST00000160134.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr11_-_116164928 | 0.34 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr2_+_32665781 | 0.34 |
ENSMUST00000066352.6
|
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chrX_+_35592006 | 0.34 |
ENSMUST00000016383.10
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr5_-_137531413 | 0.34 |
ENSMUST00000168746.8
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr6_-_108162513 | 0.33 |
ENSMUST00000167338.8
ENSMUST00000172188.2 ENSMUST00000032191.16 |
Sumf1
|
sulfatase modifying factor 1 |
| chr14_-_55880980 | 0.33 |
ENSMUST00000132338.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr9_+_44290787 | 0.33 |
ENSMUST00000066601.13
|
Hyou1
|
hypoxia up-regulated 1 |
| chr19_-_6886965 | 0.32 |
ENSMUST00000173091.2
|
Prdx5
|
peroxiredoxin 5 |
| chr8_+_26210064 | 0.32 |
ENSMUST00000068916.16
ENSMUST00000139836.8 |
Plpp5
|
phospholipid phosphatase 5 |
| chr4_+_46489248 | 0.32 |
ENSMUST00000030018.5
|
Nans
|
N-acetylneuraminic acid synthase (sialic acid synthase) |
| chr19_-_6887361 | 0.32 |
ENSMUST00000025904.12
|
Prdx5
|
peroxiredoxin 5 |
| chr7_-_45116316 | 0.31 |
ENSMUST00000033093.10
|
Bax
|
BCL2-associated X protein |
| chr14_-_45556018 | 0.30 |
ENSMUST00000022378.9
|
Ero1a
|
endoplasmic reticulum oxidoreductase 1 alpha |
| chr9_+_108216433 | 0.30 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr8_-_86281946 | 0.30 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr12_-_13299197 | 0.30 |
ENSMUST00000071103.10
|
Ddx1
|
DEAD box helicase 1 |
| chr12_+_51424343 | 0.30 |
ENSMUST00000219434.2
ENSMUST00000021335.7 |
Scfd1
|
Sec1 family domain containing 1 |
| chr8_+_13807652 | 0.29 |
ENSMUST00000130173.9
ENSMUST00000043962.9 ENSMUST00000134645.8 |
Cdc16
|
CDC16 cell division cycle 16 |
| chr1_-_93373364 | 0.29 |
ENSMUST00000190321.7
ENSMUST00000042498.14 |
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr7_+_105289729 | 0.29 |
ENSMUST00000210350.2
ENSMUST00000151193.2 ENSMUST00000209588.2 ENSMUST00000106780.2 ENSMUST00000106784.2 ENSMUST00000106785.8 ENSMUST00000106786.8 ENSMUST00000211054.2 |
Timm10b
Gm45799
|
translocase of inner mitochondrial membrane 10B predicted gene 45799 |
| chr11_-_118306620 | 0.28 |
ENSMUST00000164927.2
|
Cant1
|
calcium activated nucleotidase 1 |
| chr13_-_100969823 | 0.27 |
ENSMUST00000225922.2
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr7_-_118132520 | 0.27 |
ENSMUST00000209146.2
ENSMUST00000098090.10 ENSMUST00000032887.4 |
Coq7
|
demethyl-Q 7 |
| chr1_+_181952302 | 0.26 |
ENSMUST00000111018.2
ENSMUST00000027792.6 |
Srp9
|
signal recognition particle 9 |
| chr8_+_106895488 | 0.26 |
ENSMUST00000034378.5
ENSMUST00000212421.2 |
Slc7a6
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 |
| chr4_+_33031342 | 0.26 |
ENSMUST00000124992.8
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr14_-_55881177 | 0.26 |
ENSMUST00000138085.2
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr10_-_67120959 | 0.26 |
ENSMUST00000159002.2
ENSMUST00000077839.13 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chr6_-_119825081 | 0.26 |
ENSMUST00000183703.8
ENSMUST00000183911.8 |
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr11_-_103588487 | 0.26 |
ENSMUST00000107013.3
|
Gosr2
|
golgi SNAP receptor complex member 2 |
| chr19_+_8718837 | 0.25 |
ENSMUST00000177373.8
ENSMUST00000010254.16 |
Stx5a
|
syntaxin 5A |
| chr7_+_45522551 | 0.25 |
ENSMUST00000211234.2
|
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr4_+_109533753 | 0.25 |
ENSMUST00000102724.5
|
Faf1
|
Fas-associated factor 1 |
| chr4_-_56802266 | 0.25 |
ENSMUST00000030140.3
|
Elp1
|
elongator complex protein 1 |
| chr5_-_145138639 | 0.25 |
ENSMUST00000162220.2
ENSMUST00000031632.9 ENSMUST00000198959.2 |
Zkscan14
|
zinc finger with KRAB and SCAN domains 14 |
| chr1_-_170002444 | 0.25 |
ENSMUST00000111351.10
ENSMUST00000027981.8 |
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr7_+_97345841 | 0.25 |
ENSMUST00000026506.5
|
Clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr19_+_8719033 | 0.24 |
ENSMUST00000176314.8
ENSMUST00000073430.14 ENSMUST00000175901.8 |
Stx5a
|
syntaxin 5A |
| chr11_-_116165024 | 0.24 |
ENSMUST00000021133.16
|
Srp68
|
signal recognition particle 68 |
| chr9_-_36678868 | 0.24 |
ENSMUST00000217599.2
ENSMUST00000120381.9 |
Stt3a
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) |
| chr1_-_170002526 | 0.23 |
ENSMUST00000111350.10
|
Uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr8_+_111760521 | 0.23 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr1_+_171173252 | 0.23 |
ENSMUST00000006579.5
|
Pfdn2
|
prefoldin 2 |
| chr7_+_101859542 | 0.23 |
ENSMUST00000140631.2
ENSMUST00000120879.8 ENSMUST00000146996.8 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr2_+_61542038 | 0.23 |
ENSMUST00000028278.14
|
Psmd14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr4_+_88012836 | 0.23 |
ENSMUST00000097992.10
|
Focad
|
focadhesin |
| chr9_-_103243039 | 0.23 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr13_-_100969878 | 0.23 |
ENSMUST00000067246.6
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr3_-_37778470 | 0.23 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr15_+_6416079 | 0.22 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr10_-_18619658 | 0.22 |
ENSMUST00000215836.2
|
Arfgef3
|
ARFGEF family member 3 |
| chr8_+_124204598 | 0.22 |
ENSMUST00000001520.13
|
Afg3l1
|
AFG3-like AAA ATPase 1 |
| chr12_+_72488625 | 0.22 |
ENSMUST00000161284.3
ENSMUST00000162159.8 |
Lrrc9
|
leucine rich repeat containing 9 |
| chr4_+_130001349 | 0.22 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr4_-_150994260 | 0.22 |
ENSMUST00000134751.8
ENSMUST00000030805.14 |
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr7_+_3706992 | 0.22 |
ENSMUST00000006496.15
ENSMUST00000108623.8 ENSMUST00000139818.2 ENSMUST00000108625.8 |
Rps9
|
ribosomal protein S9 |
| chrX_-_73416824 | 0.22 |
ENSMUST00000178691.2
ENSMUST00000114146.8 |
Ubl4a
Slc10a3
|
ubiquitin-like 4A solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr5_+_31026967 | 0.22 |
ENSMUST00000114716.4
|
Tmem214
|
transmembrane protein 214 |
| chr1_+_75145275 | 0.22 |
ENSMUST00000162768.8
ENSMUST00000160439.8 ENSMUST00000027394.12 |
Zfand2b
|
zinc finger, AN1 type domain 2B |
| chr3_+_89366632 | 0.21 |
ENSMUST00000107410.8
|
Pmvk
|
phosphomevalonate kinase |
| chr7_-_44983080 | 0.21 |
ENSMUST00000211743.2
ENSMUST00000042194.10 |
Trpm4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr9_+_59446823 | 0.21 |
ENSMUST00000026262.8
|
Hexa
|
hexosaminidase A |
| chr8_-_106692668 | 0.21 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chr10_+_61516078 | 0.21 |
ENSMUST00000220372.2
ENSMUST00000020285.10 ENSMUST00000219506.2 ENSMUST00000218474.2 |
Sar1a
|
secretion associated Ras related GTPase 1A |
| chr2_+_118943274 | 0.21 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr14_-_55881257 | 0.21 |
ENSMUST00000122358.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr17_-_75858835 | 0.20 |
ENSMUST00000234785.2
ENSMUST00000112507.4 |
Fam98a
|
family with sequence similarity 98, member A |
| chr15_+_34238174 | 0.20 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr9_-_89586960 | 0.20 |
ENSMUST00000058488.9
|
Tmed3
|
transmembrane p24 trafficking protein 3 |
| chr7_-_121666486 | 0.20 |
ENSMUST00000033159.4
|
Ears2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr11_-_20781009 | 0.19 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr17_-_37269330 | 0.19 |
ENSMUST00000113669.9
|
Polr1h
|
RNA polymerase I subunit H |
| chr8_+_106099894 | 0.19 |
ENSMUST00000160650.2
|
Plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr9_+_103940575 | 0.19 |
ENSMUST00000120854.8
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr18_+_65933586 | 0.19 |
ENSMUST00000025394.14
ENSMUST00000236847.2 ENSMUST00000153193.3 |
Sec11c
|
SEC11 homolog C, signal peptidase complex subunit |
| chr3_-_36626101 | 0.18 |
ENSMUST00000029270.10
|
Ccna2
|
cyclin A2 |
| chr10_+_41395410 | 0.18 |
ENSMUST00000019962.15
|
Cd164
|
CD164 antigen |
| chr9_-_103940247 | 0.18 |
ENSMUST00000035166.12
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr4_+_154041687 | 0.18 |
ENSMUST00000105645.9
ENSMUST00000058393.9 ENSMUST00000141493.8 |
A430005L14Rik
|
RIKEN cDNA A430005L14 gene |
| chr11_+_94520567 | 0.17 |
ENSMUST00000021239.7
|
Lrrc59
|
leucine rich repeat containing 59 |
| chr7_-_45175570 | 0.17 |
ENSMUST00000167273.2
ENSMUST00000042105.11 |
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr11_+_94544593 | 0.17 |
ENSMUST00000025278.8
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr10_-_78248771 | 0.17 |
ENSMUST00000062678.11
|
Rrp1
|
ribosomal RNA processing 1 |
| chr18_-_84703738 | 0.17 |
ENSMUST00000025546.17
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr4_-_101122433 | 0.17 |
ENSMUST00000149297.2
ENSMUST00000102781.10 |
Jak1
|
Janus kinase 1 |
| chr2_+_23046381 | 0.16 |
ENSMUST00000028117.4
|
Yme1l1
|
YME1-like 1 (S. cerevisiae) |
| chr5_-_148989821 | 0.16 |
ENSMUST00000139443.8
ENSMUST00000085546.13 |
Hmgb1
|
high mobility group box 1 |
| chr8_-_34614187 | 0.16 |
ENSMUST00000033910.9
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr10_-_128759817 | 0.15 |
ENSMUST00000131271.2
|
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr11_-_60702081 | 0.15 |
ENSMUST00000018744.15
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr1_+_75119809 | 0.15 |
ENSMUST00000186037.7
ENSMUST00000187901.2 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr3_-_96812610 | 0.15 |
ENSMUST00000029738.14
|
Gpr89
|
G protein-coupled receptor 89 |
| chr5_+_107479023 | 0.15 |
ENSMUST00000031215.15
ENSMUST00000112677.10 |
Brdt
|
bromodomain, testis-specific |
| chrX_-_73416869 | 0.15 |
ENSMUST00000073067.11
ENSMUST00000037967.6 |
Slc10a3
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 3 |
| chr5_+_129970882 | 0.14 |
ENSMUST00000201855.2
ENSMUST00000073945.6 |
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr6_-_119825020 | 0.14 |
ENSMUST00000184864.8
|
Erc1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr7_+_127511681 | 0.14 |
ENSMUST00000033070.9
|
Kat8
|
K(lysine) acetyltransferase 8 |
| chr16_-_13720915 | 0.14 |
ENSMUST00000115803.9
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr14_-_55880708 | 0.14 |
ENSMUST00000120041.8
ENSMUST00000121937.8 ENSMUST00000133707.2 ENSMUST00000002391.15 ENSMUST00000121791.8 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr18_+_31742565 | 0.14 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr9_+_43996236 | 0.14 |
ENSMUST00000065461.9
ENSMUST00000176416.8 |
Usp2
|
ubiquitin specific peptidase 2 |
| chr12_+_69230931 | 0.14 |
ENSMUST00000060579.10
|
Mgat2
|
mannoside acetylglucosaminyltransferase 2 |
| chr16_-_94171533 | 0.14 |
ENSMUST00000113910.8
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_+_71819534 | 0.14 |
ENSMUST00000110054.8
ENSMUST00000139541.8 |
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr2_+_164587948 | 0.14 |
ENSMUST00000109327.4
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr4_-_133728168 | 0.14 |
ENSMUST00000105887.8
ENSMUST00000012262.12 ENSMUST00000144668.8 ENSMUST00000105889.4 |
Dhdds
|
dehydrodolichyl diphosphate synthase |
| chr10_+_128158328 | 0.13 |
ENSMUST00000219037.2
ENSMUST00000026446.4 |
Cnpy2
|
canopy FGF signaling regulator 2 |
| chrX_+_106192510 | 0.13 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr17_-_37269425 | 0.13 |
ENSMUST00000172518.8
|
Polr1h
|
RNA polymerase I subunit H |
| chr12_+_4132567 | 0.13 |
ENSMUST00000020986.15
ENSMUST00000049584.6 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr8_+_71819492 | 0.13 |
ENSMUST00000110053.9
|
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr7_+_45434755 | 0.12 |
ENSMUST00000233503.2
ENSMUST00000120005.10 ENSMUST00000211609.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr19_-_53026965 | 0.12 |
ENSMUST00000183274.8
ENSMUST00000182097.2 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chrX_+_106193167 | 0.12 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr3_-_154798982 | 0.12 |
ENSMUST00000066568.6
|
Fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr1_+_75119419 | 0.12 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr4_+_33031433 | 0.12 |
ENSMUST00000029944.13
|
Ube2j1
|
ubiquitin-conjugating enzyme E2J 1 |
| chr5_-_137531471 | 0.12 |
ENSMUST00000143495.8
ENSMUST00000111020.8 ENSMUST00000111023.8 ENSMUST00000111038.8 |
Gnb2
Epo
|
guanine nucleotide binding protein (G protein), beta 2 erythropoietin |
| chr2_+_163916042 | 0.12 |
ENSMUST00000018353.14
|
Stk4
|
serine/threonine kinase 4 |
| chr17_+_45874800 | 0.11 |
ENSMUST00000224905.2
ENSMUST00000226086.2 ENSMUST00000041353.7 |
Slc35b2
|
solute carrier family 35, member B2 |
| chrX_+_106193060 | 0.11 |
ENSMUST00000125676.8
ENSMUST00000180182.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr14_-_59835285 | 0.11 |
ENSMUST00000022555.11
ENSMUST00000225839.2 ENSMUST00000056997.15 ENSMUST00000171683.3 ENSMUST00000167100.9 |
Cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr3_+_123061094 | 0.11 |
ENSMUST00000047923.12
ENSMUST00000200333.2 |
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr10_-_18619439 | 0.11 |
ENSMUST00000019999.7
|
Arfgef3
|
ARFGEF family member 3 |
| chrX_+_73356597 | 0.11 |
ENSMUST00000114160.2
|
Fam50a
|
family with sequence similarity 50, member A |
| chr7_-_79974166 | 0.11 |
ENSMUST00000047362.11
ENSMUST00000121882.8 |
Rccd1
|
RCC1 domain containing 1 |
| chr9_-_64928927 | 0.11 |
ENSMUST00000036615.7
|
Hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr6_+_29348068 | 0.11 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr1_+_185095232 | 0.11 |
ENSMUST00000046514.13
|
Eprs
|
glutamyl-prolyl-tRNA synthetase |
| chr13_+_22129246 | 0.10 |
ENSMUST00000176511.8
ENSMUST00000102978.8 ENSMUST00000152258.9 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr12_+_111644528 | 0.10 |
ENSMUST00000221089.2
ENSMUST00000084947.10 ENSMUST00000168338.2 ENSMUST00000222737.2 |
Trmt61a
|
tRNA methyltransferase 61A |
| chr5_-_148931957 | 0.10 |
ENSMUST00000147473.6
|
Gm42791
|
predicted gene 42791 |
| chr9_-_108903117 | 0.10 |
ENSMUST00000161521.8
ENSMUST00000045011.9 |
Atrip
|
ATR interacting protein |
| chr13_+_76246853 | 0.10 |
ENSMUST00000091466.4
ENSMUST00000224386.2 |
Ttc37
|
tetratricopeptide repeat domain 37 |
| chr11_-_96807273 | 0.10 |
ENSMUST00000103152.11
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr11_+_119989736 | 0.10 |
ENSMUST00000106223.4
ENSMUST00000185558.2 |
Ndufaf8
|
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
| chr10_+_41395870 | 0.09 |
ENSMUST00000189300.2
|
Cd164
|
CD164 antigen |
| chr17_+_87415049 | 0.09 |
ENSMUST00000041369.8
ENSMUST00000234803.2 |
Socs5
|
suppressor of cytokine signaling 5 |
| chr2_+_155118217 | 0.09 |
ENSMUST00000029128.4
|
Map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr8_+_84682136 | 0.09 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr9_+_14695933 | 0.09 |
ENSMUST00000034405.11
ENSMUST00000115632.10 ENSMUST00000147305.2 |
Mre11a
|
MRE11A homolog A, double strand break repair nuclease |
| chr7_-_105289515 | 0.09 |
ENSMUST00000133519.8
ENSMUST00000209550.2 ENSMUST00000210911.2 ENSMUST00000084782.10 ENSMUST00000131446.8 |
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr10_-_128758757 | 0.09 |
ENSMUST00000135161.2
|
Rdh5
|
retinol dehydrogenase 5 |
| chr10_+_77458109 | 0.09 |
ENSMUST00000174510.8
ENSMUST00000172813.2 |
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr18_+_57487800 | 0.08 |
ENSMUST00000025490.10
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr3_-_108469468 | 0.08 |
ENSMUST00000106622.3
|
Tmem167b
|
transmembrane protein 167B |
| chr5_-_100186728 | 0.08 |
ENSMUST00000153442.8
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr11_+_69871952 | 0.08 |
ENSMUST00000108593.8
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1 |
| chr9_+_108216466 | 0.08 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr12_-_87519032 | 0.08 |
ENSMUST00000021428.9
|
Snw1
|
SNW domain containing 1 |
| chr12_+_98594388 | 0.08 |
ENSMUST00000048402.12
ENSMUST00000101144.10 ENSMUST00000101146.4 |
Spata7
|
spermatogenesis associated 7 |
| chr10_+_126877798 | 0.08 |
ENSMUST00000006915.14
ENSMUST00000120542.8 |
Mettl1
|
methyltransferase like 1 |
| chr19_+_8718777 | 0.08 |
ENSMUST00000176381.8
|
Stx5a
|
syntaxin 5A |
| chr7_+_105289655 | 0.07 |
ENSMUST00000058333.10
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr13_-_104953370 | 0.07 |
ENSMUST00000022228.13
|
Cwc27
|
CWC27 spliceosome-associated protein |
| chr13_-_38712387 | 0.07 |
ENSMUST00000035988.16
|
Txndc5
|
thioredoxin domain containing 5 |
| chr8_+_95328694 | 0.07 |
ENSMUST00000211983.2
|
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr11_-_96807233 | 0.07 |
ENSMUST00000130774.2
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr4_-_40269778 | 0.07 |
ENSMUST00000042575.7
|
Topors
|
topoisomerase I binding, arginine/serine-rich |
| chr7_-_79973863 | 0.07 |
ENSMUST00000123189.2
|
Rccd1
|
RCC1 domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0032976 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.3 | 1.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.3 | 1.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 0.6 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.4 | GO:1990773 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.1 | 0.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.4 | GO:0010269 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.1 | 0.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.9 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 1.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.5 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.7 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.2 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.1 | 0.2 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.1 | 0.3 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.7 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.2 | GO:0018323 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) glycolate biosynthetic process(GO:0046295) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.2 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:1990441 | negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990441) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.7 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.4 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.4 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 1.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 1.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 0.0 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.2 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0060300 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0097144 | BAX complex(GO:0097144) |
| 0.1 | 1.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 0.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.3 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.5 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 1.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.8 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0031597 | proteasome regulatory particle, lid subcomplex(GO:0008541) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.2 | 1.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.2 | 0.5 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.9 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 1.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.3 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.2 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.2 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.9 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |