Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
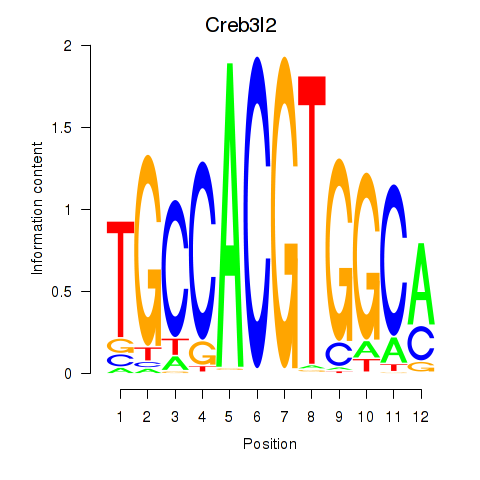
Results for Creb3l2
Z-value: 0.98
Transcription factors associated with Creb3l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb3l2
|
ENSMUSG00000038648.6 | cAMP responsive element binding protein 3-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l2 | mm39_v1_chr6_-_37419030_37419117 | -0.94 | 1.8e-02 | Click! |
Activity profile of Creb3l2 motif
Sorted Z-values of Creb3l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_128745214 | 1.00 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr3_+_104545974 | 0.73 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chrX_+_138464065 | 0.71 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr7_-_79765042 | 0.66 |
ENSMUST00000206714.2
ENSMUST00000107384.10 |
Idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr14_-_56499690 | 0.65 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr16_-_78173645 | 0.51 |
ENSMUST00000023570.14
|
Btg3
|
BTG anti-proliferation factor 3 |
| chr17_-_12988492 | 0.48 |
ENSMUST00000024599.14
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr16_-_95928804 | 0.47 |
ENSMUST00000233292.2
ENSMUST00000050884.16 |
Hmgn1
|
high mobility group nucleosomal binding domain 1 |
| chr6_-_83294526 | 0.47 |
ENSMUST00000005810.9
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr10_+_125802084 | 0.44 |
ENSMUST00000074807.8
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr5_+_31212165 | 0.43 |
ENSMUST00000202795.4
ENSMUST00000201182.4 ENSMUST00000200953.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr10_+_61484331 | 0.40 |
ENSMUST00000020286.7
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr5_+_31212110 | 0.39 |
ENSMUST00000013773.12
ENSMUST00000201838.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr3_-_101511971 | 0.38 |
ENSMUST00000036493.8
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr7_+_97345841 | 0.36 |
ENSMUST00000026506.5
|
Clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr8_+_111760521 | 0.36 |
ENSMUST00000034441.8
|
Aars
|
alanyl-tRNA synthetase |
| chr10_+_80008768 | 0.34 |
ENSMUST00000041882.7
|
Fam174c
|
family with sequence similarity 174, member C |
| chrX_+_49930311 | 0.34 |
ENSMUST00000114887.9
|
Stk26
|
serine/threonine kinase 26 |
| chr15_-_97665524 | 0.34 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr4_-_149858694 | 0.33 |
ENSMUST00000105686.3
|
Slc25a33
|
solute carrier family 25, member 33 |
| chr17_-_29566774 | 0.32 |
ENSMUST00000095427.12
ENSMUST00000118366.9 |
Mtch1
|
mitochondrial carrier 1 |
| chr14_-_55880980 | 0.31 |
ENSMUST00000132338.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr17_-_24382638 | 0.30 |
ENSMUST00000129523.3
ENSMUST00000138685.3 ENSMUST00000040735.12 |
Amdhd2
|
amidohydrolase domain containing 2 |
| chr13_-_94422337 | 0.29 |
ENSMUST00000022197.15
ENSMUST00000152555.8 |
Scamp1
|
secretory carrier membrane protein 1 |
| chr15_+_34495441 | 0.28 |
ENSMUST00000052290.14
ENSMUST00000079028.6 |
Pop1
|
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae) |
| chr19_+_6135013 | 0.28 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr6_-_22356098 | 0.27 |
ENSMUST00000165576.8
|
Fam3c
|
family with sequence similarity 3, member C |
| chr17_+_29251602 | 0.26 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr2_+_130116357 | 0.25 |
ENSMUST00000136621.9
ENSMUST00000141872.2 |
Nop56
|
NOP56 ribonucleoprotein |
| chr14_-_55881177 | 0.25 |
ENSMUST00000138085.2
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr19_-_40576782 | 0.25 |
ENSMUST00000176939.8
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr17_+_87415049 | 0.25 |
ENSMUST00000041369.8
ENSMUST00000234803.2 |
Socs5
|
suppressor of cytokine signaling 5 |
| chr6_-_13871454 | 0.24 |
ENSMUST00000185219.2
ENSMUST00000185361.2 ENSMUST00000155856.2 |
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr14_-_70680882 | 0.24 |
ENSMUST00000000793.13
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr10_-_67120959 | 0.24 |
ENSMUST00000159002.2
ENSMUST00000077839.13 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chr19_-_40576817 | 0.24 |
ENSMUST00000175932.2
ENSMUST00000176955.8 ENSMUST00000149476.3 |
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr9_+_4376556 | 0.24 |
ENSMUST00000212075.2
|
Msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chrX_-_135642025 | 0.23 |
ENSMUST00000155207.8
ENSMUST00000080411.13 ENSMUST00000169418.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr7_-_45116316 | 0.23 |
ENSMUST00000033093.10
|
Bax
|
BCL2-associated X protein |
| chr11_+_20151406 | 0.23 |
ENSMUST00000020358.12
ENSMUST00000109602.8 ENSMUST00000109601.8 ENSMUST00000163483.2 |
Rab1a
|
RAB1A, member RAS oncogene family |
| chr10_-_119075910 | 0.23 |
ENSMUST00000020315.13
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr7_+_105289729 | 0.22 |
ENSMUST00000210350.2
ENSMUST00000151193.2 ENSMUST00000209588.2 ENSMUST00000106780.2 ENSMUST00000106784.2 ENSMUST00000106785.8 ENSMUST00000106786.8 ENSMUST00000211054.2 |
Timm10b
Gm45799
|
translocase of inner mitochondrial membrane 10B predicted gene 45799 |
| chr19_-_40576897 | 0.22 |
ENSMUST00000025979.13
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr15_-_97665679 | 0.22 |
ENSMUST00000129223.9
ENSMUST00000126854.9 ENSMUST00000135080.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr2_+_129040677 | 0.20 |
ENSMUST00000028880.10
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr5_+_143389573 | 0.19 |
ENSMUST00000110731.4
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chrX_-_135641869 | 0.19 |
ENSMUST00000166930.8
ENSMUST00000113095.8 |
Morf4l2
|
mortality factor 4 like 2 |
| chr10_+_61516078 | 0.19 |
ENSMUST00000220372.2
ENSMUST00000020285.10 ENSMUST00000219506.2 ENSMUST00000218474.2 |
Sar1a
|
secretion associated Ras related GTPase 1A |
| chr4_-_119279551 | 0.19 |
ENSMUST00000106316.2
ENSMUST00000030385.13 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr14_-_55881257 | 0.18 |
ENSMUST00000122358.8
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr14_-_70680659 | 0.17 |
ENSMUST00000180358.3
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr6_-_22356175 | 0.17 |
ENSMUST00000081288.14
|
Fam3c
|
family with sequence similarity 3, member C |
| chr11_+_100960838 | 0.17 |
ENSMUST00000001802.10
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| chr13_+_90237824 | 0.17 |
ENSMUST00000012566.9
|
Tmem167
|
transmembrane protein 167 |
| chr4_+_109533753 | 0.17 |
ENSMUST00000102724.5
|
Faf1
|
Fas-associated factor 1 |
| chr2_-_164675357 | 0.16 |
ENSMUST00000042775.5
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr2_+_180367056 | 0.16 |
ENSMUST00000094218.4
|
Slc17a9
|
solute carrier family 17, member 9 |
| chr3_-_63872189 | 0.16 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr18_+_65933586 | 0.15 |
ENSMUST00000025394.14
ENSMUST00000236847.2 ENSMUST00000153193.3 |
Sec11c
|
SEC11 homolog C, signal peptidase complex subunit |
| chr13_-_55469735 | 0.15 |
ENSMUST00000035242.9
|
Rab24
|
RAB24, member RAS oncogene family |
| chr11_-_60702081 | 0.15 |
ENSMUST00000018744.15
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr5_+_31026967 | 0.15 |
ENSMUST00000114716.4
|
Tmem214
|
transmembrane protein 214 |
| chr12_-_25147139 | 0.15 |
ENSMUST00000221761.2
|
Id2
|
inhibitor of DNA binding 2 |
| chr11_-_95966407 | 0.15 |
ENSMUST00000107686.8
ENSMUST00000107684.2 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr15_-_75550581 | 0.15 |
ENSMUST00000000958.9
|
Top1mt
|
DNA topoisomerase 1, mitochondrial |
| chr1_+_171173252 | 0.15 |
ENSMUST00000006579.5
|
Pfdn2
|
prefoldin 2 |
| chr9_-_43151179 | 0.15 |
ENSMUST00000034512.7
|
Oaf
|
out at first homolog |
| chr7_-_105289515 | 0.14 |
ENSMUST00000133519.8
ENSMUST00000209550.2 ENSMUST00000210911.2 ENSMUST00000084782.10 ENSMUST00000131446.8 |
Arfip2
|
ADP-ribosylation factor interacting protein 2 |
| chr5_+_107479023 | 0.14 |
ENSMUST00000031215.15
ENSMUST00000112677.10 |
Brdt
|
bromodomain, testis-specific |
| chr6_-_22356067 | 0.14 |
ENSMUST00000163963.8
|
Fam3c
|
family with sequence similarity 3, member C |
| chr3_-_37778470 | 0.14 |
ENSMUST00000108105.2
ENSMUST00000079755.5 ENSMUST00000099128.2 |
Gm5148
|
predicted gene 5148 |
| chr12_+_17594795 | 0.13 |
ENSMUST00000171737.3
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr6_-_13871475 | 0.13 |
ENSMUST00000139231.2
|
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr9_+_108216433 | 0.13 |
ENSMUST00000191997.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr11_+_94544593 | 0.13 |
ENSMUST00000025278.8
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr2_-_32584132 | 0.13 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr1_+_75119419 | 0.12 |
ENSMUST00000097694.11
ENSMUST00000190240.7 |
Retreg2
|
reticulophagy regulator family member 2 |
| chr14_-_55880708 | 0.12 |
ENSMUST00000120041.8
ENSMUST00000121937.8 ENSMUST00000133707.2 ENSMUST00000002391.15 ENSMUST00000121791.8 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr6_-_145021816 | 0.12 |
ENSMUST00000111742.8
ENSMUST00000048252.11 |
Bcat1
|
branched chain aminotransferase 1, cytosolic |
| chr2_+_69691906 | 0.12 |
ENSMUST00000090852.11
ENSMUST00000166411.8 |
Ssb
|
Sjogren syndrome antigen B |
| chr14_-_70588803 | 0.12 |
ENSMUST00000143153.2
ENSMUST00000127000.2 ENSMUST00000068044.14 ENSMUST00000022688.10 |
Slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr10_+_81220252 | 0.12 |
ENSMUST00000072751.13
|
Dohh
|
deoxyhypusine hydroxylase/monooxygenase |
| chr19_+_34899811 | 0.11 |
ENSMUST00000223776.2
|
Kif20b
|
kinesin family member 20B |
| chr7_-_29869126 | 0.11 |
ENSMUST00000062181.9
|
Zfp146
|
zinc finger protein 146 |
| chr19_-_6817955 | 0.10 |
ENSMUST00000239527.1
|
RPS6KA4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr11_+_5049404 | 0.10 |
ENSMUST00000134267.8
ENSMUST00000036320.12 ENSMUST00000150632.8 |
Rhbdd3
|
rhomboid domain containing 3 |
| chr2_+_69691982 | 0.10 |
ENSMUST00000112260.3
|
Ssb
|
Sjogren syndrome antigen B |
| chr14_+_58063668 | 0.10 |
ENSMUST00000022538.5
|
Mrpl57
|
mitochondrial ribosomal protein L57 |
| chr3_-_63872079 | 0.10 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr15_-_34495329 | 0.10 |
ENSMUST00000022946.6
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chr3_+_89680867 | 0.09 |
ENSMUST00000038356.13
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr5_-_25047577 | 0.08 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr2_+_164675697 | 0.08 |
ENSMUST00000143780.9
|
Ctsa
|
cathepsin A |
| chr10_+_20024203 | 0.08 |
ENSMUST00000020173.16
|
Map7
|
microtubule-associated protein 7 |
| chr7_+_105289655 | 0.08 |
ENSMUST00000058333.10
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chrX_-_8011952 | 0.08 |
ENSMUST00000115615.9
ENSMUST00000115616.8 ENSMUST00000115621.9 |
Rbm3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr19_+_46561801 | 0.08 |
ENSMUST00000026011.8
|
Sfxn2
|
sideroflexin 2 |
| chr15_-_102044658 | 0.07 |
ENSMUST00000154032.2
|
Spryd3
|
SPRY domain containing 3 |
| chr6_-_8778439 | 0.07 |
ENSMUST00000115520.8
ENSMUST00000038403.12 ENSMUST00000115518.8 |
Ica1
|
islet cell autoantigen 1 |
| chr8_+_95328694 | 0.07 |
ENSMUST00000211983.2
|
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr19_+_8848876 | 0.06 |
ENSMUST00000166407.9
|
Ubxn1
|
UBX domain protein 1 |
| chr8_+_95328558 | 0.06 |
ENSMUST00000060389.10
ENSMUST00000212729.2 ENSMUST00000121101.2 |
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr2_+_132688558 | 0.06 |
ENSMUST00000028835.13
ENSMUST00000110122.10 |
Crls1
|
cardiolipin synthase 1 |
| chr7_+_79042052 | 0.05 |
ENSMUST00000118959.8
ENSMUST00000036865.13 |
Fanci
|
Fanconi anemia, complementation group I |
| chrX_+_73372664 | 0.05 |
ENSMUST00000004326.4
|
Plxna3
|
plexin A3 |
| chr5_-_35836761 | 0.05 |
ENSMUST00000114233.3
|
Htra3
|
HtrA serine peptidase 3 |
| chr5_-_135280063 | 0.04 |
ENSMUST00000062572.3
|
Fzd9
|
frizzled class receptor 9 |
| chr1_-_163552693 | 0.04 |
ENSMUST00000159679.8
|
Mettl11b
|
methyltransferase like 11B |
| chr6_-_8778106 | 0.04 |
ENSMUST00000151758.2
ENSMUST00000115519.8 ENSMUST00000153390.8 |
Ica1
|
islet cell autoantigen 1 |
| chrX_+_151922936 | 0.04 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr2_-_35869636 | 0.04 |
ENSMUST00000028248.11
ENSMUST00000112976.9 |
Ttll11
|
tubulin tyrosine ligase-like family, member 11 |
| chr9_+_7571397 | 0.04 |
ENSMUST00000120900.8
ENSMUST00000151853.8 ENSMUST00000152878.3 |
Mmp27
|
matrix metallopeptidase 27 |
| chr18_+_75500600 | 0.04 |
ENSMUST00000026999.10
|
Smad7
|
SMAD family member 7 |
| chr2_+_150628655 | 0.04 |
ENSMUST00000045441.8
|
Pygb
|
brain glycogen phosphorylase |
| chr3_-_107993906 | 0.04 |
ENSMUST00000102638.8
ENSMUST00000102637.8 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr16_-_5021843 | 0.03 |
ENSMUST00000147567.2
ENSMUST00000023911.11 |
Nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr7_-_33896033 | 0.03 |
ENSMUST00000148381.3
ENSMUST00000002710.10 |
Pdcd2l
|
programmed cell death 2-like |
| chr1_+_63216281 | 0.03 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr7_-_24950145 | 0.03 |
ENSMUST00000116343.3
ENSMUST00000045847.15 |
Erf
|
Ets2 repressor factor |
| chr5_-_31311496 | 0.03 |
ENSMUST00000201353.4
ENSMUST00000200864.4 ENSMUST00000200833.4 ENSMUST00000201491.2 ENSMUST00000200744.4 ENSMUST00000202241.4 ENSMUST00000154241.8 |
Mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr18_-_40352372 | 0.03 |
ENSMUST00000025364.6
|
Yipf5
|
Yip1 domain family, member 5 |
| chr2_-_92201342 | 0.02 |
ENSMUST00000176810.8
ENSMUST00000090582.11 ENSMUST00000068586.13 |
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr11_-_120534469 | 0.02 |
ENSMUST00000141254.8
ENSMUST00000170556.8 ENSMUST00000151876.8 ENSMUST00000026133.15 ENSMUST00000139706.2 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr8_+_14961702 | 0.02 |
ENSMUST00000161162.8
ENSMUST00000110800.9 |
Arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr11_+_100436433 | 0.02 |
ENSMUST00000092684.12
ENSMUST00000006976.8 |
Odad4
|
outer dynein arm complex subunit 4 |
| chr6_-_77956635 | 0.02 |
ENSMUST00000161846.8
ENSMUST00000160894.8 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr9_+_108216466 | 0.02 |
ENSMUST00000193987.2
|
Gpx1
|
glutathione peroxidase 1 |
| chr2_+_71283620 | 0.01 |
ENSMUST00000037210.9
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr2_-_126718037 | 0.01 |
ENSMUST00000028843.12
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr8_+_14961663 | 0.01 |
ENSMUST00000084207.12
|
Arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr8_-_106863521 | 0.01 |
ENSMUST00000115979.9
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr15_-_83234372 | 0.01 |
ENSMUST00000226124.2
ENSMUST00000067215.9 |
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr16_+_20551853 | 0.01 |
ENSMUST00000115423.8
ENSMUST00000007171.13 ENSMUST00000232646.2 |
Chrd
|
chordin |
| chr2_-_162929732 | 0.01 |
ENSMUST00000094653.6
|
Gtsf1l
|
gametocyte specific factor 1-like |
| chr6_-_93769426 | 0.01 |
ENSMUST00000204788.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr4_+_104623505 | 0.01 |
ENSMUST00000031663.10
ENSMUST00000065072.7 |
C8b
|
complement component 8, beta polypeptide |
| chr4_-_108263873 | 0.01 |
ENSMUST00000184609.2
|
Gpx7
|
glutathione peroxidase 7 |
| chr9_-_67739607 | 0.01 |
ENSMUST00000054500.7
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr10_+_20024724 | 0.01 |
ENSMUST00000116259.5
ENSMUST00000214231.2 |
Map7
|
microtubule-associated protein 7 |
| chr15_+_77617804 | 0.01 |
ENSMUST00000230377.2
|
Apol9b
|
apolipoprotein L 9b |
| chr11_-_103844870 | 0.01 |
ENSMUST00000103075.11
|
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr11_+_109253598 | 0.01 |
ENSMUST00000106702.4
ENSMUST00000020930.14 |
Gna13
|
guanine nucleotide binding protein, alpha 13 |
| chr2_-_92201311 | 0.01 |
ENSMUST00000176774.8
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr13_-_24390265 | 0.01 |
ENSMUST00000123076.8
|
Carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr6_+_87864796 | 0.01 |
ENSMUST00000113607.10
ENSMUST00000049966.6 |
Copg1
|
coatomer protein complex, subunit gamma 1 |
| chr7_+_32535979 | 0.01 |
ENSMUST00000189118.2
|
Scgb1b15
|
secretoglobin, family 1B, member 15 |
| chr7_+_32762874 | 0.01 |
ENSMUST00000187081.2
|
Scgb1b17
|
secretoglobin, family 1B, member 17 |
| chr8_-_95328348 | 0.00 |
ENSMUST00000212547.2
ENSMUST00000212507.2 ENSMUST00000034226.8 |
Psme3ip1
|
proteasome activator subunit 3 interacting protein 1 |
| chrX_+_156601431 | 0.00 |
ENSMUST00000087157.5
|
Klhl34
|
kelch-like 34 |
| chrX_+_8137372 | 0.00 |
ENSMUST00000127103.8
ENSMUST00000115591.8 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr16_-_30369378 | 0.00 |
ENSMUST00000140402.8
|
Tmem44
|
transmembrane protein 44 |
| chr2_+_153716958 | 0.00 |
ENSMUST00000028983.3
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr4_+_109835224 | 0.00 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr5_-_137608726 | 0.00 |
ENSMUST00000197912.5
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chrX_-_133377225 | 0.00 |
ENSMUST00000009740.9
|
Taf7l
|
TATA-box binding protein associated factor 7 like |
| chr5_-_139799953 | 0.00 |
ENSMUST00000044002.10
|
Tmem184a
|
transmembrane protein 184a |
| chr6_-_40917431 | 0.00 |
ENSMUST00000122181.8
ENSMUST00000031935.10 |
1700074P13Rik
|
RIKEN cDNA 1700074P13 gene |
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb3l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.2 | 0.7 | GO:1990773 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.2 | 1.0 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 0.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.1 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.6 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 | 0.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.4 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.8 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.2 | GO:1990117 | release of matrix enzymes from mitochondria(GO:0032976) B cell receptor apoptotic signaling pathway(GO:1990117) |
| 0.1 | 0.4 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 0.3 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.3 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.3 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0010269 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) response to selenium ion(GO:0010269) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.4 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.6 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.1 | 0.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.2 | GO:0097144 | BAX complex(GO:0097144) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 0.7 | GO:0019202 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.4 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.0 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |