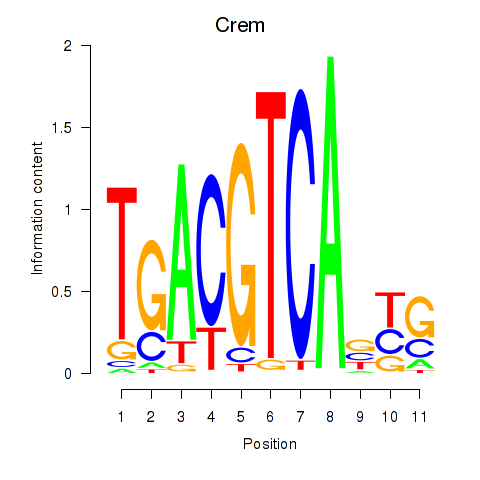
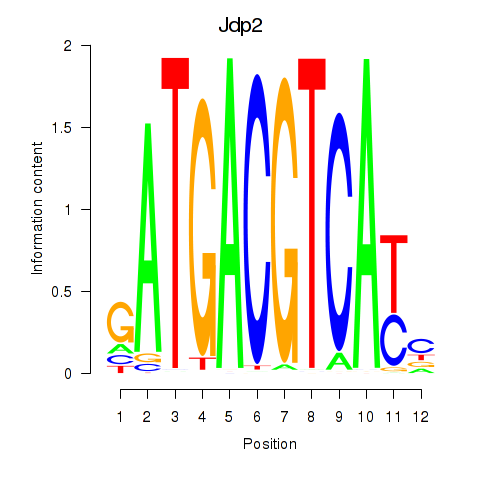
chr4_-_133972890
Show fit
0.79
ENSMUST00000030644.8
Zfp593
zinc finger protein 593
chr6_-_124441731
Show fit
0.61
ENSMUST00000008297.5
Clstn3
calsyntenin 3
chr11_+_52123016
Show fit
0.49
ENSMUST00000109072.2
Skp1
S-phase kinase-associated protein 1
chr7_-_30428746
Show fit
0.48
ENSMUST00000209065.2
ENSMUST00000208169.2
Tmem147
transmembrane protein 147
chr11_-_76134436
Show fit
0.42
ENSMUST00000164022.8
ENSMUST00000168055.2
ENSMUST00000169701.8
Glod4
glyoxalase domain containing 4
chr12_+_52144511
Show fit
0.41
ENSMUST00000040090.16
Nubpl
nucleotide binding protein-like
chr8_-_106738514
Show fit
0.41
ENSMUST00000058579.7
Ddx28
DEAD box helicase 28
chr7_-_30428930
Show fit
0.40
ENSMUST00000207296.2
ENSMUST00000006478.10
Tmem147
transmembrane protein 147
chr11_+_76134541
Show fit
0.40
ENSMUST00000040577.5
Mrm3
mitochondrial rRNA methyltransferase 3
chr4_+_141028539
Show fit
0.38
ENSMUST00000006614.3
Epha2
Eph receptor A2
chr17_-_28126679
Show fit
0.36
ENSMUST00000025057.5
Taf11
TATA-box binding protein associated factor 11
chr9_+_106048116
Show fit
0.36
ENSMUST00000020490.13
Wdr82
WD repeat domain containing 82
chr16_-_3895642
Show fit
0.35
ENSMUST00000006137.9
Trap1
TNF receptor-associated protein 1
chr11_+_52122836
Show fit
0.34
ENSMUST00000037324.12
ENSMUST00000166537.8
Skp1
S-phase kinase-associated protein 1
chr5_-_137529251
Show fit
0.34
ENSMUST00000132525.8
Gnb2
guanine nucleotide binding protein (G protein), beta 2
chr11_-_76134513
Show fit
0.34
ENSMUST00000017430.12
Glod4
glyoxalase domain containing 4
chr8_+_106738105
Show fit
0.33
ENSMUST00000034375.11
Dus2
dihydrouridine synthase 2
chr19_-_4675352
Show fit
0.33
ENSMUST00000224707.2
ENSMUST00000237059.2
Rce1
Ras converting CAAX endopeptidase 1
chr5_-_121329385
Show fit
0.33
ENSMUST00000054547.9
ENSMUST00000100770.9
Ptpn11
protein tyrosine phosphatase, non-receptor type 11
chr18_+_35347983
Show fit
0.33
ENSMUST00000235449.2
ENSMUST00000235269.2
Ctnna1
catenin (cadherin associated protein), alpha 1
chr9_-_57375269
Show fit
0.32
ENSMUST00000215059.2
ENSMUST00000046587.8
ENSMUST00000214256.2
Scamp5
secretory carrier membrane protein 5
chr13_-_111626562
Show fit
0.30
ENSMUST00000091236.11
ENSMUST00000047627.14
Gpbp1
GC-rich promoter binding protein 1
chr19_-_4675631
Show fit
0.30
ENSMUST00000225375.2
ENSMUST00000025823.6
Rce1
Ras converting CAAX endopeptidase 1
chr4_-_123644091
Show fit
0.30
ENSMUST00000102636.4
Akirin1
akirin 1
chr4_+_11704438
Show fit
0.29
ENSMUST00000108304.9
Gem
GTP binding protein (gene overexpressed in skeletal muscle)
chr6_+_17749169
Show fit
0.29
ENSMUST00000053148.14
ENSMUST00000115417.4
St7
suppression of tumorigenicity 7
chr7_-_30364394
Show fit
0.28
ENSMUST00000019697.9
Haus5
HAUS augmin-like complex, subunit 5
chr19_-_4675300
Show fit
0.28
ENSMUST00000225264.2
ENSMUST00000237022.2
ENSMUST00000224675.3
Rce1
Ras converting CAAX endopeptidase 1
chr11_+_83553400
Show fit
0.28
ENSMUST00000019074.4
Ccl4
chemokine (C-C motif) ligand 4
chr3_-_107603778
Show fit
0.27
ENSMUST00000029490.15
Ahcyl1
S-adenosylhomocysteine hydrolase-like 1
chr15_+_34238174
Show fit
0.26
ENSMUST00000022867.5
ENSMUST00000226627.2
Laptm4b
lysosomal-associated protein transmembrane 4B
chr10_+_79977291
Show fit
0.26
ENSMUST00000105367.8
Atp5d
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit
chr17_-_31731222
Show fit
0.26
ENSMUST00000236665.2
Wdr4
WD repeat domain 4
chr4_-_94940425
Show fit
0.26
ENSMUST00000107094.2
Jun
jun proto-oncogene
chr12_+_72807985
Show fit
0.26
ENSMUST00000021514.10
Ppm1a
protein phosphatase 1A, magnesium dependent, alpha isoform
chr9_-_119151428
Show fit
0.25
ENSMUST00000040853.11
Oxsr1
oxidative-stress responsive 1
chr12_-_102709884
Show fit
0.23
ENSMUST00000173760.9
ENSMUST00000178384.2
Moap1
modulator of apoptosis 1
chr7_-_19043955
Show fit
0.23
ENSMUST00000207334.2
ENSMUST00000208505.2
ENSMUST00000207716.2
ENSMUST00000208326.2
ENSMUST00000003640.4
Fosb
FBJ osteosarcoma oncogene B
chr3_+_144824325
Show fit
0.23
ENSMUST00000098538.9
ENSMUST00000106192.9
ENSMUST00000098539.7
ENSMUST00000029920.15
Odf2l
outer dense fiber of sperm tails 2-like
chr17_+_47201552
Show fit
0.23
ENSMUST00000040434.9
Tbcc
tubulin-specific chaperone C
chr6_-_83030759
Show fit
0.23
ENSMUST00000134606.8
Htra2
HtrA serine peptidase 2
chr2_+_36026395
Show fit
0.22
ENSMUST00000028250.9
Mrrf
mitochondrial ribosome recycling factor
chr9_-_96360648
Show fit
0.22
ENSMUST00000071301.5
Rnf7
ring finger protein 7
chr9_+_20914211
Show fit
0.22
ENSMUST00000214124.2
ENSMUST00000216818.2
Mrpl4
mitochondrial ribosomal protein L4
chr5_-_137529465
Show fit
0.22
ENSMUST00000150063.9
Gnb2
guanine nucleotide binding protein (G protein), beta 2
chr4_-_56802266
Show fit
0.22
ENSMUST00000030140.3
Elp1
elongator complex protein 1
chr8_+_124204598
Show fit
0.22
ENSMUST00000001520.13
Afg3l1
AFG3-like AAA ATPase 1
chr11_-_70128712
Show fit
0.21
ENSMUST00000108577.8
ENSMUST00000108579.8
ENSMUST00000021181.13
ENSMUST00000108578.9
ENSMUST00000102569.10
0610010K14Rik
RIKEN cDNA 0610010K14 gene
chr7_+_90091937
Show fit
0.21
ENSMUST00000061767.5
Crebzf
CREB/ATF bZIP transcription factor
chr6_-_50543514
Show fit
0.21
ENSMUST00000161401.2
Cycs
cytochrome c, somatic
chr17_+_36290743
Show fit
0.20
ENSMUST00000087200.4
Gnl1
guanine nucleotide binding protein-like 1
chr12_+_4132567
Show fit
0.20
ENSMUST00000020986.15
ENSMUST00000049584.6
Dnajc27
DnaJ heat shock protein family (Hsp40) member C27
chr9_-_64080161
Show fit
0.20
ENSMUST00000176299.8
ENSMUST00000130127.8
ENSMUST00000176794.8
ENSMUST00000177045.8
Zwilch
zwilch kinetochore protein
chr8_-_61407760
Show fit
0.20
ENSMUST00000110302.8
Clcn3
chloride channel, voltage-sensitive 3
chr11_+_93935021
Show fit
0.20
ENSMUST00000075695.13
ENSMUST00000092777.11
Spag9
sperm associated antigen 9
chr2_+_164587948
Show fit
0.19
ENSMUST00000109327.4
Dnttip1
deoxynucleotidyltransferase, terminal, interacting protein 1
chr11_+_93935066
Show fit
0.19
ENSMUST00000103168.10
Spag9
sperm associated antigen 9
chr10_+_128173603
Show fit
0.19
ENSMUST00000005826.9
Cs
citrate synthase
chr1_+_75145275
Show fit
0.19
ENSMUST00000162768.8
ENSMUST00000160439.8
ENSMUST00000027394.12
Zfand2b
Links
zinc finger, AN1 type domain 2B
chr9_-_96360676
Show fit
0.18
ENSMUST00000057500.6
Rnf7
ring finger protein 7
chr9_+_113641615
Show fit
0.18
ENSMUST00000111838.10
ENSMUST00000166734.10
ENSMUST00000214522.2
ENSMUST00000163895.3
Clasp2
CLIP associating protein 2
chr11_-_70128462
Show fit
0.18
ENSMUST00000100950.10
0610010K14Rik
RIKEN cDNA 0610010K14 gene
chr12_-_110662256
Show fit
0.18
ENSMUST00000149189.2
Hsp90aa1
heat shock protein 90, alpha (cytosolic), class A member 1
chr8_+_121395047
Show fit
0.18
ENSMUST00000181795.2
Cox4i1
cytochrome c oxidase subunit 4I1
chr12_+_75355082
Show fit
0.18
ENSMUST00000118602.8
ENSMUST00000118966.8
ENSMUST00000055390.6
Rhoj
ras homolog family member J
chr12_+_108758871
Show fit
0.17
ENSMUST00000021692.9
Yy1
YY1 transcription factor
chr10_-_17823736
Show fit
0.17
ENSMUST00000037879.8
Heca
hdc homolog, cell cycle regulator
chr6_+_125016723
Show fit
0.17
ENSMUST00000140131.8
ENSMUST00000032480.14
Ing4
inhibitor of growth family, member 4
chr19_-_24938909
Show fit
0.17
ENSMUST00000025815.10
Cbwd1
COBW domain containing 1
chr10_-_117681864
Show fit
0.17
ENSMUST00000064667.9
Rap1b
RAS related protein 1b
chr14_-_123220554
Show fit
0.16
ENSMUST00000126867.8
ENSMUST00000148661.2
ENSMUST00000037726.14
Tmtc4
transmembrane and tetratricopeptide repeat containing 4
chr5_+_114582327
Show fit
0.16
ENSMUST00000137167.8
ENSMUST00000112239.9
ENSMUST00000124260.8
ENSMUST00000125650.6
ENSMUST00000043760.15
Mvk
mevalonate kinase
chr12_-_86931529
Show fit
0.16
ENSMUST00000038422.8
Irf2bpl
interferon regulatory factor 2 binding protein-like
chr12_+_84363603
Show fit
0.16
ENSMUST00000045931.12
Zfp410
zinc finger protein 410
chr17_+_29879569
Show fit
0.16
ENSMUST00000024816.13
ENSMUST00000235031.2
ENSMUST00000234911.2
Cmtr1
cap methyltransferase 1
chr7_+_101859542
Show fit
0.16
ENSMUST00000140631.2
ENSMUST00000120879.8
ENSMUST00000146996.8
Pgap2
post-GPI attachment to proteins 2
chr11_-_88608920
Show fit
0.16
ENSMUST00000092794.12
Msi2
musashi RNA-binding protein 2
chr19_+_11747721
Show fit
0.16
ENSMUST00000167199.3
Mrpl16
mitochondrial ribosomal protein L16
chr1_+_75213044
Show fit
0.16
ENSMUST00000188931.7
Dnajb2
DnaJ heat shock protein family (Hsp40) member B2
chr2_+_180098026
Show fit
0.16
ENSMUST00000038259.13
Slco4a1
solute carrier organic anion transporter family, member 4a1
chr7_+_3706992
Show fit
0.15
ENSMUST00000006496.15
ENSMUST00000108623.8
ENSMUST00000139818.2
ENSMUST00000108625.8
Rps9
ribosomal protein S9
chr18_+_14916295
Show fit
0.15
ENSMUST00000234789.2
ENSMUST00000169862.2
Taf4b
TATA-box binding protein associated factor 4b
chrX_-_55643429
Show fit
0.15
ENSMUST00000059899.3
Mmgt1
membrane magnesium transporter 1
chr3_+_134918298
Show fit
0.15
ENSMUST00000062893.12
Cenpe
centromere protein E
chr17_+_29879684
Show fit
0.15
ENSMUST00000235014.2
ENSMUST00000130423.4
Cmtr1
cap methyltransferase 1
chr4_+_130001349
Show fit
0.15
ENSMUST00000030563.6
Pef1
penta-EF hand domain containing 1
chr7_+_97437709
Show fit
0.15
ENSMUST00000206984.2
Pak1
p21 (RAC1) activated kinase 1
chr2_-_36026614
Show fit
0.15
ENSMUST00000122456.8
Rbm18
RNA binding motif protein 18
chr19_-_42117420
Show fit
0.15
ENSMUST00000161873.2
ENSMUST00000018965.4
Avpi1
arginine vasopressin-induced 1
chr6_-_83031358
Show fit
0.15
ENSMUST00000113962.8
ENSMUST00000089645.13
ENSMUST00000113963.8
Htra2
HtrA serine peptidase 2
chr5_-_93354287
Show fit
0.14
ENSMUST00000144514.3
Ccni
cyclin I
chr8_+_121394961
Show fit
0.14
ENSMUST00000034276.13
ENSMUST00000181586.8
Cox4i1
cytochrome c oxidase subunit 4I1
chr4_-_126215462
Show fit
0.14
ENSMUST00000102617.5
Adprs
ADP-ribosylserine hydrolase
chr7_+_125202653
Show fit
0.14
ENSMUST00000206103.2
ENSMUST00000033000.8
Il21r
interleukin 21 receptor
chr7_-_118132520
Show fit
0.14
ENSMUST00000209146.2
ENSMUST00000098090.10
ENSMUST00000032887.4
Coq7
demethyl-Q 7
chr11_-_70128678
Show fit
0.14
ENSMUST00000108575.9
0610010K14Rik
RIKEN cDNA 0610010K14 gene
chr6_-_131270136
Show fit
0.14
ENSMUST00000032307.12
Magohb
mago homolog B, exon junction complex core component
chr18_+_38552011
Show fit
0.14
ENSMUST00000025293.5
Ndfip1
Nedd4 family interacting protein 1
chr10_+_81012465
Show fit
0.14
ENSMUST00000047864.11
Eef2
eukaryotic translation elongation factor 2
chr8_-_70078152
Show fit
0.13
ENSMUST00000121886.8
Zfp868
zinc finger protein 868
chr11_+_93935156
Show fit
0.13
ENSMUST00000024979.15
Spag9
sperm associated antigen 9
chr5_+_143534455
Show fit
0.13
ENSMUST00000169329.8
ENSMUST00000067145.12
ENSMUST00000119488.2
ENSMUST00000118121.2
ENSMUST00000200267.2
ENSMUST00000196487.2
Fam220a
Fam220a
family with sequence similarity 220, member A
chr5_+_33978035
Show fit
0.13
ENSMUST00000075812.11
ENSMUST00000114397.9
ENSMUST00000155880.8
Nsd2
nuclear receptor binding SET domain protein 2
chr10_+_44144346
Show fit
0.13
ENSMUST00000039286.5
Atg5
autophagy related 5
chr4_-_129556234
Show fit
0.13
ENSMUST00000003828.11
Kpna6
karyopherin (importin) alpha 6
chr10_+_72490705
Show fit
0.13
ENSMUST00000105431.8
ENSMUST00000160337.2
Zwint
ZW10 interactor
chr2_-_36026785
Show fit
0.13
ENSMUST00000028251.10
Rbm18
RNA binding motif protein 18
chr13_-_19579898
Show fit
0.13
ENSMUST00000197565.3
ENSMUST00000221380.2
ENSMUST00000200323.3
ENSMUST00000199924.2
ENSMUST00000222869.2
Stard3nl
STARD3 N-terminal like
chr7_+_138448308
Show fit
0.13
ENSMUST00000155672.8
Ppp2r2d
protein phosphatase 2, regulatory subunit B, delta
chr16_+_49519561
Show fit
0.13
ENSMUST00000046777.11
ENSMUST00000142682.9
Ift57
intraflagellar transport 57
chr12_+_112586501
Show fit
0.13
ENSMUST00000180015.9
Adssl1
adenylosuccinate synthetase like 1
chr8_+_3671599
Show fit
0.13
ENSMUST00000207389.2
Pet100
PET100 homolog
chr14_+_70815250
Show fit
0.13
ENSMUST00000228554.2
Nudt18
nudix (nucleoside diphosphate linked moiety X)-type motif 18
chr1_+_172327812
Show fit
0.13
ENSMUST00000192460.2
Tagln2
transgelin 2
chr9_+_64080644
Show fit
0.13
ENSMUST00000034966.9
Rpl4
ribosomal protein L4
chr12_-_69205882
Show fit
0.13
ENSMUST00000037023.9
Rps29
ribosomal protein S29
chr1_-_92401459
Show fit
0.13
ENSMUST00000185251.2
ENSMUST00000027478.7
Ndufa10
NADH:ubiquinone oxidoreductase subunit A10
chr17_-_37269153
Show fit
0.12
ENSMUST00000172823.2
Polr1h
RNA polymerase I subunit H
chr6_+_129510117
Show fit
0.12
ENSMUST00000032264.9
Gabarapl1
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1
chr3_-_62414391
Show fit
0.12
ENSMUST00000029336.6
Dhx36
DEAH (Asp-Glu-Ala-His) box polypeptide 36
chr2_+_155907100
Show fit
0.12
ENSMUST00000038860.12
Spag4
sperm associated antigen 4
chr15_+_65682066
Show fit
0.12
ENSMUST00000211878.2
Efr3a
EFR3 homolog A
chr9_+_108437485
Show fit
0.12
ENSMUST00000081111.14
ENSMUST00000193421.2
Impdh2
inosine monophosphate dehydrogenase 2
chr9_+_77824646
Show fit
0.12
ENSMUST00000034904.14
Elovl5
ELOVL family member 5, elongation of long chain fatty acids (yeast)
chr11_-_79145489
Show fit
0.12
ENSMUST00000017821.12
Wsb1
WD repeat and SOCS box-containing 1
chr18_-_80194682
Show fit
0.12
ENSMUST00000066743.11
Adnp2
ADNP homeobox 2
chr11_+_52287239
Show fit
0.12
ENSMUST00000036952.5
ENSMUST00000109057.8
9530068E07Rik
RIKEN cDNA 9530068E07 gene
chr16_-_92196954
Show fit
0.12
ENSMUST00000023672.10
Rcan1
regulator of calcineurin 1
chr15_-_79571977
Show fit
0.12
ENSMUST00000023061.7
Josd1
Josephin domain containing 1
chr5_-_114582053
Show fit
0.12
ENSMUST00000123256.3
ENSMUST00000112245.6
Mmab
methylmalonic aciduria (cobalamin deficiency) cblB type homolog (human)
chr2_-_32314017
Show fit
0.11
ENSMUST00000113307.9
Slc25a25
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25
chr17_+_84819260
Show fit
0.11
ENSMUST00000047206.7
Plekhh2
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
chr9_+_66620959
Show fit
0.11
ENSMUST00000071889.13
Car12
carbonic anhydrase 12
chr7_+_138448061
Show fit
0.11
ENSMUST00000041097.13
Ppp2r2d
protein phosphatase 2, regulatory subunit B, delta
chr15_+_99291455
Show fit
0.11
ENSMUST00000162624.8
Tmbim6
transmembrane BAX inhibitor motif containing 6
chr9_+_69305180
Show fit
0.11
ENSMUST00000034761.15
ENSMUST00000125938.2
Ice2
interactor of little elongation complex ELL subunit 2
chr2_+_120439858
Show fit
0.11
ENSMUST00000124187.8
Haus2
HAUS augmin-like complex, subunit 2
chr11_-_70128587
Show fit
0.11
ENSMUST00000108576.10
0610010K14Rik
RIKEN cDNA 0610010K14 gene
chr9_+_66621001
Show fit
0.11
ENSMUST00000085420.12
Car12
carbonic anhydrase 12
chr5_+_123214332
Show fit
0.11
ENSMUST00000067505.15
ENSMUST00000111619.10
ENSMUST00000160344.2
Tmem120b
transmembrane protein 120B
chr8_-_80438260
Show fit
0.11
ENSMUST00000080536.8
Abce1
ATP-binding cassette, sub-family E (OABP), member 1
chr7_-_7301760
Show fit
0.11
ENSMUST00000210061.2
Clcn4
chloride channel, voltage-sensitive 4
chr7_+_46495256
Show fit
0.11
ENSMUST00000048209.16
ENSMUST00000210815.2
ENSMUST00000125862.8
ENSMUST00000210968.2
ENSMUST00000092621.12
ENSMUST00000210467.2
Ldha
lactate dehydrogenase A
chr1_-_59276252
Show fit
0.11
ENSMUST00000163058.2
ENSMUST00000160945.2
ENSMUST00000027178.13
Als2
alsin Rho guanine nucleotide exchange factor
chr12_+_112586465
Show fit
0.10
ENSMUST00000021726.8
Adssl1
adenylosuccinate synthetase like 1
chr17_-_31731190
Show fit
0.10
ENSMUST00000237127.2
Wdr4
WD repeat domain 4
chr7_+_46495521
Show fit
0.10
ENSMUST00000133062.2
Ldha
lactate dehydrogenase A
chr2_+_32665781
Show fit
0.10
ENSMUST00000066352.6
Ptrh1
peptidyl-tRNA hydrolase 1 homolog
chr13_-_19579961
Show fit
0.10
ENSMUST00000039694.13
Stard3nl
STARD3 N-terminal like
chr7_-_97768725
Show fit
0.10
ENSMUST00000107128.8
Myo7a
myosin VIIA
chr8_+_78276020
Show fit
0.10
ENSMUST00000056237.15
ENSMUST00000118622.2
Prmt9
protein arginine methyltransferase 9
chr7_-_4504635
Show fit
0.10
ENSMUST00000013886.9
Ppp1r12c
protein phosphatase 1, regulatory subunit 12C
chr16_-_32065972
Show fit
0.10
ENSMUST00000042732.6
Fbxo45
F-box protein 45
chr10_-_128383508
Show fit
0.10
ENSMUST00000152539.8
ENSMUST00000133458.2
ENSMUST00000040572.10
Zc3h10
zinc finger CCCH type containing 10
chr18_+_66591604
Show fit
0.10
ENSMUST00000025399.9
ENSMUST00000237161.2
ENSMUST00000236933.2
Pmaip1
phorbol-12-myristate-13-acetate-induced protein 1
chr10_+_80465481
Show fit
0.10
ENSMUST00000085435.7
Csnk1g2
casein kinase 1, gamma 2
chr17_-_36291087
Show fit
0.10
ENSMUST00000055454.14
Prr3
proline-rich polypeptide 3
chr3_-_157742204
Show fit
0.10
ENSMUST00000137444.8
Srsf11
serine and arginine-rich splicing factor 11
chr9_-_59393893
Show fit
0.10
ENSMUST00000171975.8
Arih1
ariadne RBR E3 ubiquitin protein ligase 1
chr6_+_83162928
Show fit
0.10
ENSMUST00000113907.2
Dctn1
dynactin 1
chrX_-_165992145
Show fit
0.09
ENSMUST00000112176.8
Tmsb4x
thymosin, beta 4, X chromosome
chr10_+_72490677
Show fit
0.09
ENSMUST00000020081.11
Zwint
ZW10 interactor
chr10_+_81019076
Show fit
0.09
ENSMUST00000219133.2
ENSMUST00000047665.7
Dapk3
death-associated protein kinase 3
chr2_+_164298854
Show fit
0.09
ENSMUST00000109352.8
Sys1
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae)
chr2_+_162773440
Show fit
0.09
ENSMUST00000130411.7
ENSMUST00000126163.3
Srsf6
serine and arginine-rich splicing factor 6
chr1_+_75213114
Show fit
0.09
ENSMUST00000188290.7
Dnajb2
DnaJ heat shock protein family (Hsp40) member B2
chrX_+_65696608
Show fit
0.09
ENSMUST00000036043.5
Slitrk2
SLIT and NTRK-like family, member 2
chr17_-_80203457
Show fit
0.09
ENSMUST00000068282.7
ENSMUST00000112437.8
Atl2
atlastin GTPase 2
chr9_-_45866264
Show fit
0.09
ENSMUST00000114573.9
Sidt2
SID1 transmembrane family, member 2
chr10_+_81019117
Show fit
0.09
ENSMUST00000218157.2
Dapk3
death-associated protein kinase 3
chr2_-_118987305
Show fit
0.09
ENSMUST00000135419.8
ENSMUST00000129351.2
ENSMUST00000139519.2
ENSMUST00000094695.12
Rmdn3
regulator of microtubule dynamics 3
chr1_-_36748985
Show fit
0.09
ENSMUST00000043951.10
Actr1b
ARP1 actin-related protein 1B, centractin beta
chr11_+_93934940
Show fit
0.09
ENSMUST00000132079.8
Spag9
sperm associated antigen 9
chr5_+_24369961
Show fit
0.09
ENSMUST00000049887.13
Nupl2
nucleoporin like 2
chr8_+_106428256
Show fit
0.09
ENSMUST00000093195.7
ENSMUST00000211888.2
ENSMUST00000212430.2
Pard6a
par-6 family cell polarity regulator alpha
chr1_-_134289670
Show fit
0.09
ENSMUST00000049470.11
Tmem183a
transmembrane protein 183A
chr11_+_102992508
Show fit
0.09
ENSMUST00000107040.10
ENSMUST00000140372.8
ENSMUST00000024492.15
ENSMUST00000134884.8
Acbd4
acyl-Coenzyme A binding domain containing 4
chr1_-_165064455
Show fit
0.09
ENSMUST00000043235.8
Tiprl
TIP41, TOR signalling pathway regulator-like (S. cerevisiae)
chr11_+_97692738
Show fit
0.09
ENSMUST00000127033.9
Lasp1
LIM and SH3 protein 1
chr14_-_59835285
Show fit
0.09
ENSMUST00000022555.11
ENSMUST00000225839.2
ENSMUST00000056997.15
ENSMUST00000171683.3
ENSMUST00000167100.9
Cdadc1
cytidine and dCMP deaminase domain containing 1
chr1_+_75213082
Show fit
0.09
ENSMUST00000055223.14
ENSMUST00000082158.13
ENSMUST00000188346.7
Dnajb2
DnaJ heat shock protein family (Hsp40) member B2
chr13_+_120151982
Show fit
0.09
ENSMUST00000179869.3
ENSMUST00000224188.2
Hmgcs1
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1
chr5_-_4154681
Show fit
0.09
ENSMUST00000001507.5
Cyp51
cytochrome P450, family 51
chr11_+_101556367
Show fit
0.08
ENSMUST00000039388.3
Arl4d
ADP-ribosylation factor-like 4D
chr3_+_40754489
Show fit
0.08
ENSMUST00000203295.3
Plk4
polo like kinase 4
chr8_+_89423645
Show fit
0.08
ENSMUST00000043526.15
ENSMUST00000211554.2
ENSMUST00000209532.2
ENSMUST00000209559.2
Cyld
CYLD lysine 63 deubiquitinase
chr8_+_123502236
Show fit
0.08
ENSMUST00000015160.6
ENSMUST00000212781.2
ENSMUST00000212790.2
Acsf3
acyl-CoA synthetase family member 3
chr11_+_22940599
Show fit
0.08
ENSMUST00000020562.5
Cct4
chaperonin containing Tcp1, subunit 4 (delta)
chr11_-_90281721
Show fit
0.08
ENSMUST00000004051.8
Hlf
hepatic leukemia factor
chr17_-_26288447
Show fit
0.08
ENSMUST00000122103.9
ENSMUST00000120691.9
Rab11fip3
RAB11 family interacting protein 3 (class II)
chr3_+_31149201
Show fit
0.08
ENSMUST00000118470.8
ENSMUST00000029194.12
ENSMUST00000123532.2
Skil
SKI-like
chr17_-_71833752
Show fit
0.08
ENSMUST00000232863.2
ENSMUST00000024851.10
Ndc80
NDC80 kinetochore complex component
chr2_-_120439981
Show fit
0.08
ENSMUST00000133612.2
ENSMUST00000102498.8
ENSMUST00000102499.8
Lrrc57
leucine rich repeat containing 57
chr8_+_11777721
Show fit
0.08
ENSMUST00000210104.2
Arhgef7
Rho guanine nucleotide exchange factor (GEF7)
chr7_-_118183892
Show fit
0.08
ENSMUST00000044195.6
Tmc7
transmembrane channel-like gene family 7
chr13_+_93440265
Show fit
0.08
ENSMUST00000109494.8
Homer1
homer scaffolding protein 1
chr11_-_87295292
Show fit
0.08
ENSMUST00000067692.13
Rad51c
RAD51 paralog C
chr17_-_45884179
Show fit
0.08
ENSMUST00000165127.8
ENSMUST00000166469.8
ENSMUST00000024739.14
Hsp90ab1
heat shock protein 90 alpha (cytosolic), class B member 1
chr5_-_147662798
Show fit
0.07
ENSMUST00000110529.6
ENSMUST00000031653.12
Flt1
FMS-like tyrosine kinase 1
chr17_+_32255071
Show fit
0.07
ENSMUST00000081339.13
Rrp1b
ribosomal RNA processing 1B