Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Crx_Gsc
Z-value: 0.14
Transcription factors associated with Crx_Gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
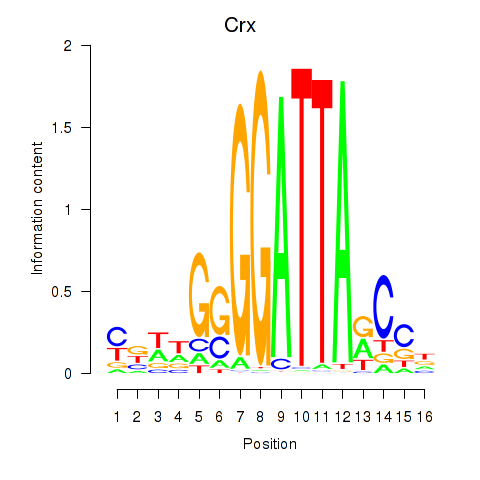
Crx
|
ENSMUSG00000041578.16 | cone-rod homeobox |
|
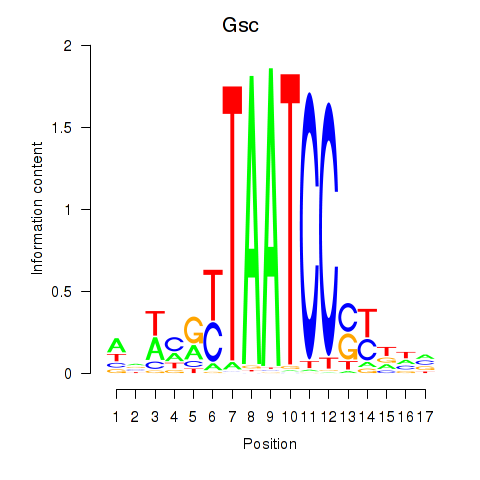
Gsc
|
ENSMUSG00000021095.6 | goosecoid homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gsc | mm39_v1_chr12_-_104439589_104439633 | -0.65 | 2.3e-01 | Click! |
| Crx | mm39_v1_chr7_-_15613769_15613893 | -0.43 | 4.6e-01 | Click! |
Activity profile of Crx_Gsc motif
Sorted Z-values of Crx_Gsc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_101943134 | 0.18 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr11_-_53321242 | 0.12 |
ENSMUST00000109019.8
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr2_+_3771709 | 0.10 |
ENSMUST00000177037.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chrM_+_7758 | 0.08 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr11_+_109540201 | 0.08 |
ENSMUST00000106677.8
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha |
| chr8_-_71326027 | 0.08 |
ENSMUST00000212680.2
|
Ccdc124
|
coiled-coil domain containing 124 |
| chr10_-_93727003 | 0.07 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr14_-_20027112 | 0.07 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr10_-_75617245 | 0.07 |
ENSMUST00000001715.10
|
Gstt3
|
glutathione S-transferase, theta 3 |
| chr14_+_51193449 | 0.06 |
ENSMUST00000095925.5
|
Pnp2
|
purine-nucleoside phosphorylase 2 |
| chr11_-_116165024 | 0.06 |
ENSMUST00000021133.16
|
Srp68
|
signal recognition particle 68 |
| chr11_+_29668563 | 0.06 |
ENSMUST00000060992.6
|
Rtn4
|
reticulon 4 |
| chr1_-_149798090 | 0.06 |
ENSMUST00000111926.9
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr19_-_4976844 | 0.06 |
ENSMUST00000236496.2
|
Dpp3
|
dipeptidylpeptidase 3 |
| chr10_+_79977291 | 0.06 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr10_+_55982969 | 0.05 |
ENSMUST00000063138.8
|
Msl3l2
|
MSL3 like 2 |
| chr12_-_13299136 | 0.05 |
ENSMUST00000221623.2
|
Ddx1
|
DEAD box helicase 1 |
| chr9_+_88209250 | 0.05 |
ENSMUST00000034992.8
|
Nt5e
|
5' nucleotidase, ecto |
| chr11_+_90078485 | 0.05 |
ENSMUST00000124877.3
ENSMUST00000153734.2 ENSMUST00000134929.8 |
Gm45716
|
predicted gene 45716 |
| chr9_-_106768601 | 0.05 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr2_+_144435974 | 0.05 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr11_-_4045343 | 0.05 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr8_+_3671599 | 0.05 |
ENSMUST00000207389.2
|
Pet100
|
PET100 homolog |
| chr10_-_9776823 | 0.05 |
ENSMUST00000141722.8
|
Stxbp5
|
syntaxin binding protein 5 (tomosyn) |
| chrX_+_37689503 | 0.05 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr14_-_76248274 | 0.04 |
ENSMUST00000088922.5
|
Gtf2f2
|
general transcription factor IIF, polypeptide 2 |
| chr1_-_135241429 | 0.04 |
ENSMUST00000134088.3
ENSMUST00000081104.10 |
Timm17a
|
translocase of inner mitochondrial membrane 17a |
| chr7_+_65710086 | 0.04 |
ENSMUST00000153609.8
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr19_-_10859046 | 0.04 |
ENSMUST00000128835.8
|
Tmem109
|
transmembrane protein 109 |
| chr15_+_72962241 | 0.04 |
ENSMUST00000089765.9
ENSMUST00000134353.2 |
Chrac1
|
chromatin accessibility complex 1 |
| chr8_+_79236051 | 0.04 |
ENSMUST00000209992.2
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr17_+_34416707 | 0.04 |
ENSMUST00000025196.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr7_+_142606476 | 0.04 |
ENSMUST00000037941.10
|
Cd81
|
CD81 antigen |
| chr12_-_87312982 | 0.04 |
ENSMUST00000179379.9
|
Vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr11_-_116080361 | 0.04 |
ENSMUST00000148601.2
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chrX_-_101232978 | 0.04 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr10_+_59839142 | 0.04 |
ENSMUST00000164083.4
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr19_-_8858247 | 0.04 |
ENSMUST00000096253.7
|
Uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr16_+_33614378 | 0.04 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr7_+_92210348 | 0.04 |
ENSMUST00000032842.13
ENSMUST00000085017.5 |
Ccdc90b
|
coiled-coil domain containing 90B |
| chr10_-_75616788 | 0.04 |
ENSMUST00000173512.2
ENSMUST00000173537.2 |
Gm20441
Gstt3
|
predicted gene 20441 glutathione S-transferase, theta 3 |
| chr18_-_20995980 | 0.03 |
ENSMUST00000224530.2
|
Trappc8
|
trafficking protein particle complex 8 |
| chr5_-_137530214 | 0.03 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr15_+_76227695 | 0.03 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr14_+_26722319 | 0.03 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr11_-_116164928 | 0.03 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr17_+_26342474 | 0.03 |
ENSMUST00000025014.10
ENSMUST00000236166.2 ENSMUST00000127647.3 |
Mrpl28
|
mitochondrial ribosomal protein L28 |
| chr11_-_82799186 | 0.03 |
ENSMUST00000103213.10
|
Nle1
|
notchless homolog 1 |
| chr17_+_34416689 | 0.03 |
ENSMUST00000173441.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr9_+_73020708 | 0.03 |
ENSMUST00000169399.8
ENSMUST00000034738.14 |
Rsl24d1
|
ribosomal L24 domain containing 1 |
| chrM_+_7779 | 0.03 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr5_+_115373895 | 0.03 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr19_-_5416339 | 0.03 |
ENSMUST00000170010.3
|
Banf1
|
BAF nuclear assembly factor 1 |
| chr4_-_43840201 | 0.03 |
ENSMUST00000214281.2
|
Olfr157
|
olfactory receptor 157 |
| chr14_+_48683797 | 0.03 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chr19_+_8861096 | 0.03 |
ENSMUST00000187504.7
|
Lbhd1
|
LBH domain containing 1 |
| chr7_-_44635813 | 0.03 |
ENSMUST00000208829.2
ENSMUST00000207370.2 ENSMUST00000107843.11 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr11_-_48762170 | 0.03 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr9_-_60594742 | 0.03 |
ENSMUST00000114032.8
ENSMUST00000166168.8 ENSMUST00000132366.2 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr14_-_64654397 | 0.03 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chrX_-_99670174 | 0.03 |
ENSMUST00000015812.12
|
Pdzd11
|
PDZ domain containing 11 |
| chr17_-_13211374 | 0.03 |
ENSMUST00000159551.8
ENSMUST00000160781.8 |
Wtap
|
Wilms tumour 1-associating protein |
| chr6_-_113717689 | 0.03 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr3_+_142202642 | 0.03 |
ENSMUST00000090127.7
|
Gbp5
|
guanylate binding protein 5 |
| chr16_+_33614715 | 0.03 |
ENSMUST00000023520.7
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr6_+_17749169 | 0.03 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr14_-_70396859 | 0.03 |
ENSMUST00000058240.14
ENSMUST00000153871.2 |
9930012K11Rik
|
RIKEN cDNA 9930012K11 gene |
| chr6_-_85114725 | 0.03 |
ENSMUST00000174769.2
ENSMUST00000174286.3 ENSMUST00000045986.8 |
Spr
|
sepiapterin reductase |
| chr18_-_64649497 | 0.03 |
ENSMUST00000237351.2
ENSMUST00000236186.2 ENSMUST00000235325.2 |
Nars
|
asparaginyl-tRNA synthetase |
| chr6_+_123206802 | 0.02 |
ENSMUST00000112554.9
ENSMUST00000024118.11 ENSMUST00000117130.8 |
Clec4n
|
C-type lectin domain family 4, member n |
| chr16_+_91184661 | 0.02 |
ENSMUST00000139503.2
|
Ifnar2
|
interferon (alpha and beta) receptor 2 |
| chr3_-_63872189 | 0.02 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr11_+_83553400 | 0.02 |
ENSMUST00000019074.4
|
Ccl4
|
chemokine (C-C motif) ligand 4 |
| chr19_+_43741431 | 0.02 |
ENSMUST00000026199.14
|
Cutc
|
cutC copper transporter |
| chr4_+_155789246 | 0.02 |
ENSMUST00000030905.9
|
Ssu72
|
Ssu72 RNA polymerase II CTD phosphatase homolog (yeast) |
| chr8_-_95564881 | 0.02 |
ENSMUST00000034233.15
ENSMUST00000162538.9 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr18_+_84869883 | 0.02 |
ENSMUST00000163083.2
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr19_-_6178171 | 0.02 |
ENSMUST00000154601.8
ENSMUST00000138931.3 |
Snx15
|
sorting nexin 15 |
| chr2_+_90884349 | 0.02 |
ENSMUST00000067663.14
ENSMUST00000002171.14 ENSMUST00000111441.10 |
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
| chr7_-_113875261 | 0.02 |
ENSMUST00000135570.8
|
Psma1
|
proteasome subunit alpha 1 |
| chr6_+_117877272 | 0.02 |
ENSMUST00000177743.8
ENSMUST00000167182.8 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr17_+_29251602 | 0.02 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr13_-_90237631 | 0.02 |
ENSMUST00000160232.8
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr4_-_140501507 | 0.02 |
ENSMUST00000026381.7
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr18_+_24122689 | 0.02 |
ENSMUST00000074941.8
|
Zfp35
|
zinc finger protein 35 |
| chr7_+_133311062 | 0.02 |
ENSMUST00000033282.5
|
Bccip
|
BRCA2 and CDKN1A interacting protein |
| chr1_-_185061525 | 0.02 |
ENSMUST00000027921.11
ENSMUST00000110975.8 ENSMUST00000110974.4 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr7_-_113875342 | 0.02 |
ENSMUST00000033008.10
|
Psma1
|
proteasome subunit alpha 1 |
| chr17_+_35827997 | 0.02 |
ENSMUST00000164242.9
ENSMUST00000045956.14 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chr11_+_5470652 | 0.02 |
ENSMUST00000063084.16
|
Xbp1
|
X-box binding protein 1 |
| chr7_+_28524627 | 0.02 |
ENSMUST00000066264.13
|
Ech1
|
enoyl coenzyme A hydratase 1, peroxisomal |
| chr13_-_4659120 | 0.02 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr10_+_61516078 | 0.02 |
ENSMUST00000220372.2
ENSMUST00000020285.10 ENSMUST00000219506.2 ENSMUST00000218474.2 |
Sar1a
|
secretion associated Ras related GTPase 1A |
| chr2_+_152468850 | 0.02 |
ENSMUST00000000369.4
ENSMUST00000150913.2 |
Rem1
|
rad and gem related GTP binding protein 1 |
| chr4_-_116851571 | 0.02 |
ENSMUST00000030446.15
|
Urod
|
uroporphyrinogen decarboxylase |
| chr12_-_101942172 | 0.02 |
ENSMUST00000221191.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr11_+_95925711 | 0.02 |
ENSMUST00000006217.10
ENSMUST00000107700.4 |
Snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr19_+_5416769 | 0.02 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr12_-_118162652 | 0.02 |
ENSMUST00000084806.7
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chr17_-_87573294 | 0.02 |
ENSMUST00000145895.8
ENSMUST00000129616.8 ENSMUST00000155904.2 ENSMUST00000151155.8 ENSMUST00000144236.9 ENSMUST00000024963.11 |
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chrY_+_1010543 | 0.02 |
ENSMUST00000091197.4
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr11_-_120489172 | 0.02 |
ENSMUST00000026125.3
|
Alyref
|
Aly/REF export factor |
| chr10_-_128640232 | 0.02 |
ENSMUST00000051011.14
|
Tmem198b
|
transmembrane protein 198b |
| chr11_-_69873343 | 0.02 |
ENSMUST00000147437.3
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr12_-_79239022 | 0.02 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr7_-_44635740 | 0.02 |
ENSMUST00000209056.3
ENSMUST00000209124.2 ENSMUST00000208312.2 ENSMUST00000207659.2 ENSMUST00000045325.14 |
Prmt1
|
protein arginine N-methyltransferase 1 |
| chr16_-_94327689 | 0.02 |
ENSMUST00000023615.7
|
Vps26c
|
VPS26 endosomal protein sorting factor C |
| chr12_-_87312994 | 0.02 |
ENSMUST00000072744.15
|
Vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr18_-_74340842 | 0.02 |
ENSMUST00000040188.16
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr2_-_144112444 | 0.02 |
ENSMUST00000028909.5
|
Snx5
|
sorting nexin 5 |
| chr4_-_149822480 | 0.02 |
ENSMUST00000105687.9
ENSMUST00000054459.11 ENSMUST00000103208.2 |
Tmem201
|
transmembrane protein 201 |
| chr15_+_80017315 | 0.02 |
ENSMUST00000023050.9
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr14_+_56640038 | 0.02 |
ENSMUST00000095793.3
ENSMUST00000223627.2 |
Rnf17
|
ring finger protein 17 |
| chr9_+_38630317 | 0.02 |
ENSMUST00000129598.2
|
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr17_+_34092811 | 0.02 |
ENSMUST00000226885.2
|
Smim40-ps
|
small integral membrane protein 40, pseudogene |
| chr4_+_65523223 | 0.02 |
ENSMUST00000050850.14
ENSMUST00000107366.2 |
Trim32
|
tripartite motif-containing 32 |
| chr12_+_101942222 | 0.02 |
ENSMUST00000047357.10
|
Cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr5_+_114268425 | 0.02 |
ENSMUST00000031587.13
|
Ung
|
uracil DNA glycosylase |
| chr1_+_172328768 | 0.02 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr15_+_76832681 | 0.02 |
ENSMUST00000071792.7
ENSMUST00000230274.2 |
1110038F14Rik
|
RIKEN cDNA 1110038F14 gene |
| chr15_+_76832920 | 0.02 |
ENSMUST00000229229.2
|
1110038F14Rik
|
RIKEN cDNA 1110038F14 gene |
| chrX_+_7750483 | 0.02 |
ENSMUST00000115663.10
ENSMUST00000096514.11 |
Slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member A2 |
| chr10_-_93725619 | 0.02 |
ENSMUST00000181091.8
ENSMUST00000181217.8 ENSMUST00000047910.15 ENSMUST00000180688.2 |
Metap2
|
methionine aminopeptidase 2 |
| chr13_+_54937312 | 0.02 |
ENSMUST00000163915.8
ENSMUST00000099503.10 ENSMUST00000171859.8 |
Tspan17
|
tetraspanin 17 |
| chr16_-_30900181 | 0.02 |
ENSMUST00000055389.9
|
Xxylt1
|
xyloside xylosyltransferase 1 |
| chr12_+_85645801 | 0.02 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr8_+_120071211 | 0.01 |
ENSMUST00000034300.8
ENSMUST00000212468.2 |
Hsbp1
|
heat shock factor binding protein 1 |
| chr12_+_84332006 | 0.01 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr11_+_120489358 | 0.01 |
ENSMUST00000093140.5
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr15_-_78058214 | 0.01 |
ENSMUST00000016781.8
|
Ift27
|
intraflagellar transport 27 |
| chr5_-_137529465 | 0.01 |
ENSMUST00000150063.9
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr4_-_43025756 | 0.01 |
ENSMUST00000098109.9
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr18_-_32044877 | 0.01 |
ENSMUST00000054984.8
|
Sft2d3
|
SFT2 domain containing 3 |
| chr2_+_90884612 | 0.01 |
ENSMUST00000185715.7
|
Psmc3
|
proteasome (prosome, macropain) 26S subunit, ATPase 3 |
| chr1_+_87983189 | 0.01 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr8_+_56393488 | 0.01 |
ENSMUST00000000275.10
|
Glra3
|
glycine receptor, alpha 3 subunit |
| chr13_-_35211060 | 0.01 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr12_+_98886826 | 0.01 |
ENSMUST00000085109.10
ENSMUST00000079146.13 |
Ttc8
|
tetratricopeptide repeat domain 8 |
| chr11_+_69512850 | 0.01 |
ENSMUST00000108656.9
ENSMUST00000092969.2 |
Sat2
|
spermidine/spermine N1-acetyl transferase 2 |
| chr8_+_114860375 | 0.01 |
ENSMUST00000147605.8
ENSMUST00000134593.2 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr13_+_108182941 | 0.01 |
ENSMUST00000224361.2
ENSMUST00000224390.2 |
Smim15
|
small integral membrane protein 15 |
| chr12_-_84447625 | 0.01 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr13_+_12410240 | 0.01 |
ENSMUST00000059270.10
|
Heatr1
|
HEAT repeat containing 1 |
| chr19_-_43741363 | 0.01 |
ENSMUST00000045562.6
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr14_+_70314727 | 0.01 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr5_-_74229021 | 0.01 |
ENSMUST00000119154.8
ENSMUST00000068058.14 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr8_+_95564949 | 0.01 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr11_-_86148379 | 0.01 |
ENSMUST00000132024.8
ENSMUST00000139285.8 |
Ints2
|
integrator complex subunit 2 |
| chr2_+_174126103 | 0.01 |
ENSMUST00000109095.8
ENSMUST00000180362.8 ENSMUST00000109096.9 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr7_-_139662135 | 0.01 |
ENSMUST00000211044.2
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr11_+_70350963 | 0.01 |
ENSMUST00000126105.2
|
Zmynd15
|
zinc finger, MYND-type containing 15 |
| chr8_+_71125876 | 0.01 |
ENSMUST00000034311.15
|
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_+_122348140 | 0.01 |
ENSMUST00000196187.5
ENSMUST00000100747.3 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr7_+_44034225 | 0.01 |
ENSMUST00000120262.2
|
Syt3
|
synaptotagmin III |
| chr11_+_120489247 | 0.01 |
ENSMUST00000026128.10
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chr1_+_85992341 | 0.01 |
ENSMUST00000027432.9
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr9_-_108903117 | 0.01 |
ENSMUST00000161521.8
ENSMUST00000045011.9 |
Atrip
|
ATR interacting protein |
| chr11_+_101046708 | 0.01 |
ENSMUST00000043654.10
|
Tubg2
|
tubulin, gamma 2 |
| chr11_+_3913970 | 0.01 |
ENSMUST00000109985.8
ENSMUST00000020705.5 |
Pes1
|
pescadillo ribosomal biogenesis factor 1 |
| chr3_+_85946145 | 0.01 |
ENSMUST00000238331.2
|
Sh3d19
|
SH3 domain protein D19 |
| chr14_+_66205932 | 0.01 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chr2_+_69691906 | 0.01 |
ENSMUST00000090852.11
ENSMUST00000166411.8 |
Ssb
|
Sjogren syndrome antigen B |
| chr8_+_4303067 | 0.01 |
ENSMUST00000011981.5
ENSMUST00000208316.2 |
Snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr18_-_74340885 | 0.01 |
ENSMUST00000177604.2
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr16_+_23109213 | 0.01 |
ENSMUST00000115335.2
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1 |
| chr10_-_81332147 | 0.01 |
ENSMUST00000118498.8
|
Ncln
|
nicalin |
| chr11_-_102120917 | 0.01 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr9_+_50528608 | 0.01 |
ENSMUST00000000171.15
ENSMUST00000151197.8 |
Pih1d2
|
PIH1 domain containing 2 |
| chr4_+_21727727 | 0.01 |
ENSMUST00000108240.3
|
Ccnc
|
cyclin C |
| chr12_-_101942463 | 0.01 |
ENSMUST00000221422.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr14_-_68819544 | 0.01 |
ENSMUST00000022641.9
|
Adamdec1
|
ADAM-like, decysin 1 |
| chr11_-_69470139 | 0.01 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr11_+_96355413 | 0.01 |
ENSMUST00000103154.11
ENSMUST00000100521.10 ENSMUST00000100519.11 |
Skap1
|
src family associated phosphoprotein 1 |
| chr6_-_59403264 | 0.01 |
ENSMUST00000051065.6
|
Gprin3
|
GPRIN family member 3 |
| chr8_-_80438260 | 0.01 |
ENSMUST00000080536.8
|
Abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr14_-_20596508 | 0.01 |
ENSMUST00000161989.2
|
Ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isoform |
| chr5_-_65248927 | 0.01 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr9_+_35332837 | 0.01 |
ENSMUST00000119129.10
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr7_+_24584197 | 0.01 |
ENSMUST00000156372.8
ENSMUST00000124035.2 |
Rps19
|
ribosomal protein S19 |
| chr7_+_44033520 | 0.01 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr19_-_32689687 | 0.01 |
ENSMUST00000237752.2
ENSMUST00000235412.2 |
Atad1
|
ATPase family, AAA domain containing 1 |
| chr6_+_119213468 | 0.01 |
ENSMUST00000238905.2
ENSMUST00000037434.13 |
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr16_-_17711950 | 0.01 |
ENSMUST00000155943.9
|
Dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr5_+_143450329 | 0.01 |
ENSMUST00000045593.12
|
Daglb
|
diacylglycerol lipase, beta |
| chr6_-_66537080 | 0.01 |
ENSMUST00000079584.3
ENSMUST00000227014.2 |
Vmn1r32
|
vomeronasal 1 receptor 32 |
| chr7_-_9787223 | 0.01 |
ENSMUST00000074943.9
ENSMUST00000086298.14 |
Vmn2r50
|
vomeronasal 2, receptor 50 |
| chr14_-_63781381 | 0.01 |
ENSMUST00000058679.7
|
Mtmr9
|
myotubularin related protein 9 |
| chr8_+_57908920 | 0.01 |
ENSMUST00000034023.4
|
Scrg1
|
scrapie responsive gene 1 |
| chr16_+_32066037 | 0.01 |
ENSMUST00000141820.8
ENSMUST00000178573.8 ENSMUST00000023474.4 ENSMUST00000135289.2 |
Wdr53
|
WD repeat domain 53 |
| chr18_-_73948501 | 0.01 |
ENSMUST00000025439.5
|
Me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr4_-_43025792 | 0.01 |
ENSMUST00000067481.6
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr14_-_20596580 | 0.01 |
ENSMUST00000022355.11
ENSMUST00000161445.8 ENSMUST00000159027.8 |
Ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isoform |
| chr15_+_75812801 | 0.01 |
ENSMUST00000037260.8
|
Zfp623
|
zinc finger protein 623 |
| chr11_-_29197222 | 0.01 |
ENSMUST00000020754.10
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr10_-_59452489 | 0.01 |
ENSMUST00000020312.13
|
Mcu
|
mitochondrial calcium uniporter |
| chr7_-_139662159 | 0.01 |
ENSMUST00000166758.4
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr2_-_87902174 | 0.01 |
ENSMUST00000215268.2
|
Olfr1163
|
olfactory receptor 1163 |
| chr19_+_5074070 | 0.01 |
ENSMUST00000025826.7
ENSMUST00000237371.2 ENSMUST00000235416.2 |
Slc29a2
|
solute carrier family 29 (nucleoside transporters), member 2 |
| chr8_+_71126012 | 0.01 |
ENSMUST00000146972.3
ENSMUST00000210071.2 ENSMUST00000210987.2 |
Lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_-_80597479 | 0.01 |
ENSMUST00000026818.12
ENSMUST00000117383.8 ENSMUST00000119980.2 |
Sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Crx_Gsc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0030942 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |