Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
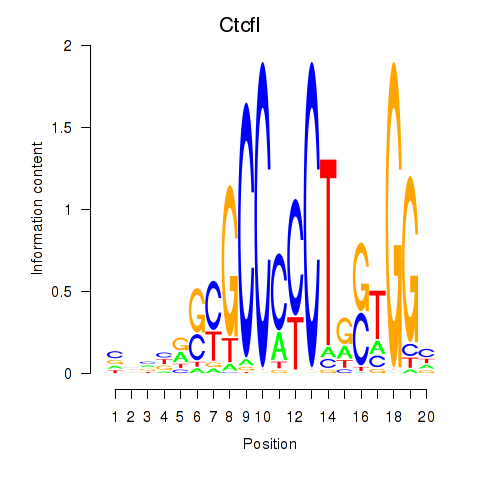
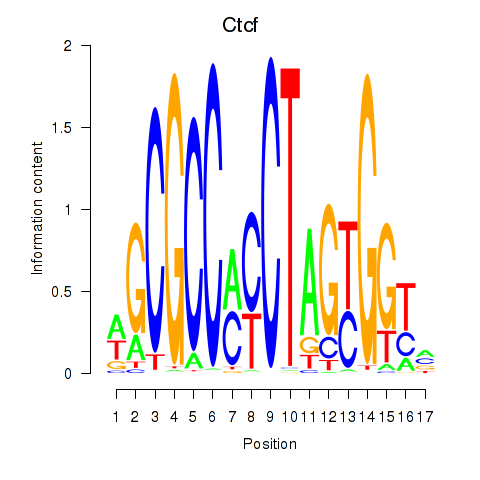
Results for Ctcfl_Ctcf
Z-value: 0.47
Transcription factors associated with Ctcfl_Ctcf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ctcfl
|
ENSMUSG00000070495.12 | CCCTC-binding factor (zinc finger protein)-like |
|
Ctcf
|
ENSMUSG00000005698.16 | CCCTC-binding factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ctcf | mm39_v1_chr8_+_106363141_106363221 | 0.94 | 1.9e-02 | Click! |
| Ctcfl | mm39_v1_chr2_-_172961168_172961318 | 0.39 | 5.1e-01 | Click! |
Activity profile of Ctcfl_Ctcf motif
Sorted Z-values of Ctcfl_Ctcf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10857734 | 0.32 |
ENSMUST00000133303.8
|
Tmem109
|
transmembrane protein 109 |
| chr14_-_14389372 | 0.30 |
ENSMUST00000023924.4
|
Rpp14
|
ribonuclease P 14 subunit |
| chr1_-_133537953 | 0.29 |
ENSMUST00000164574.2
ENSMUST00000166291.8 ENSMUST00000164096.2 ENSMUST00000166915.8 |
Snrpe
|
small nuclear ribonucleoprotein E |
| chr19_+_10160249 | 0.28 |
ENSMUST00000010807.6
|
Fads1
|
fatty acid desaturase 1 |
| chr4_-_43025756 | 0.27 |
ENSMUST00000098109.9
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr19_+_10160283 | 0.27 |
ENSMUST00000235160.2
|
Fads1
|
fatty acid desaturase 1 |
| chr15_-_36496880 | 0.26 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr10_-_126866682 | 0.25 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr4_+_140428777 | 0.23 |
ENSMUST00000138808.8
ENSMUST00000038893.6 |
Rcc2
|
regulator of chromosome condensation 2 |
| chr2_-_152857239 | 0.22 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr5_+_31078911 | 0.22 |
ENSMUST00000201571.4
|
Khk
|
ketohexokinase |
| chr17_-_56343531 | 0.22 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr19_-_6899173 | 0.20 |
ENSMUST00000025906.12
ENSMUST00000239322.2 |
Esrra
|
estrogen related receptor, alpha |
| chr4_-_43025792 | 0.20 |
ENSMUST00000067481.6
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr15_+_100659622 | 0.20 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr2_+_3285240 | 0.19 |
ENSMUST00000081932.13
|
Nmt2
|
N-myristoyltransferase 2 |
| chr6_-_124441731 | 0.18 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr4_+_152018142 | 0.18 |
ENSMUST00000062904.11
|
Dnajc11
|
DnaJ heat shock protein family (Hsp40) member C11 |
| chr12_-_110669076 | 0.17 |
ENSMUST00000155242.8
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr5_-_117254037 | 0.17 |
ENSMUST00000086471.12
|
Suds3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr4_-_126215462 | 0.17 |
ENSMUST00000102617.5
|
Adprs
|
ADP-ribosylserine hydrolase |
| chr8_+_106412905 | 0.17 |
ENSMUST00000213019.2
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr4_+_134123631 | 0.17 |
ENSMUST00000105869.9
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr16_+_16714333 | 0.16 |
ENSMUST00000027373.12
ENSMUST00000232247.2 |
Ppm1f
|
protein phosphatase 1F (PP2C domain containing) |
| chr19_-_10079091 | 0.16 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr4_+_152123772 | 0.16 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr2_+_3285269 | 0.16 |
ENSMUST00000102989.10
ENSMUST00000091504.5 |
Nmt2
|
N-myristoyltransferase 2 |
| chr1_+_16175998 | 0.16 |
ENSMUST00000027053.8
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr5_-_117254146 | 0.16 |
ENSMUST00000166397.3
|
Suds3
|
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr2_+_180224505 | 0.16 |
ENSMUST00000029085.9
|
Mrgbp
|
MRG/MORF4L binding protein |
| chr17_+_25992761 | 0.16 |
ENSMUST00000237541.2
|
Ciao3
|
cytosolic iron-sulfur assembly component 3 |
| chr2_-_66086919 | 0.16 |
ENSMUST00000125446.3
ENSMUST00000102718.10 |
Ttc21b
|
tetratricopeptide repeat domain 21B |
| chr2_+_26476619 | 0.15 |
ENSMUST00000239075.2
ENSMUST00000174211.8 ENSMUST00000145575.10 ENSMUST00000173920.9 |
Egfl7
|
EGF-like domain 7 |
| chr7_+_25016492 | 0.15 |
ENSMUST00000128119.2
|
Megf8
|
multiple EGF-like-domains 8 |
| chr8_-_108151661 | 0.15 |
ENSMUST00000003946.9
|
Nob1
|
NIN1/RPN12 binding protein 1 homolog |
| chr2_-_157988303 | 0.15 |
ENSMUST00000103122.10
|
Tgm2
|
transglutaminase 2, C polypeptide |
| chr7_+_34818709 | 0.15 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr8_-_85663976 | 0.15 |
ENSMUST00000109741.9
ENSMUST00000119820.2 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr3_-_105594865 | 0.15 |
ENSMUST00000090680.11
|
Ddx20
|
DEAD box helicase 20 |
| chr5_+_114845821 | 0.14 |
ENSMUST00000094441.11
|
Tchp
|
trichoplein, keratin filament binding |
| chr16_-_10994135 | 0.14 |
ENSMUST00000037633.16
|
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr11_+_62539657 | 0.14 |
ENSMUST00000127589.2
ENSMUST00000155759.3 |
Mmgt2
|
membrane magnesium transporter 2 |
| chr5_+_115373895 | 0.14 |
ENSMUST00000081497.13
|
Pop5
|
processing of precursor 5, ribonuclease P/MRP family (S. cerevisiae) |
| chr14_+_55842002 | 0.13 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr19_-_6899121 | 0.13 |
ENSMUST00000173635.2
|
Esrra
|
estrogen related receptor, alpha |
| chr11_-_45835737 | 0.13 |
ENSMUST00000129820.8
|
Lsm11
|
U7 snRNP-specific Sm-like protein LSM11 |
| chr3_-_107603778 | 0.13 |
ENSMUST00000029490.15
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1 |
| chr11_-_3864664 | 0.13 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr4_-_57143437 | 0.13 |
ENSMUST00000095076.10
ENSMUST00000030142.4 |
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4b |
| chr3_-_95125002 | 0.12 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr5_+_147894121 | 0.12 |
ENSMUST00000085558.11
ENSMUST00000129092.2 |
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr7_-_108769719 | 0.12 |
ENSMUST00000208136.2
ENSMUST00000036992.9 |
Lmo1
|
LIM domain only 1 |
| chr7_+_49428082 | 0.11 |
ENSMUST00000032715.13
|
Prmt3
|
protein arginine N-methyltransferase 3 |
| chr6_-_85114725 | 0.11 |
ENSMUST00000174769.2
ENSMUST00000174286.3 ENSMUST00000045986.8 |
Spr
|
sepiapterin reductase |
| chr4_-_149221998 | 0.11 |
ENSMUST00000176124.8
ENSMUST00000177408.2 ENSMUST00000105695.2 |
Cenps
|
centromere protein S |
| chr9_+_45818250 | 0.11 |
ENSMUST00000216672.2
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr11_+_4085164 | 0.11 |
ENSMUST00000003677.11
ENSMUST00000145705.8 |
Rnf215
|
ring finger protein 215 |
| chr6_+_83142902 | 0.11 |
ENSMUST00000077407.12
ENSMUST00000113913.8 ENSMUST00000130212.8 |
Dctn1
|
dynactin 1 |
| chr6_+_127423779 | 0.11 |
ENSMUST00000112191.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr8_+_107757847 | 0.11 |
ENSMUST00000034388.10
|
Vps4a
|
vacuolar protein sorting 4A |
| chr2_+_93017887 | 0.11 |
ENSMUST00000150462.8
ENSMUST00000111266.8 |
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr2_-_32602246 | 0.11 |
ENSMUST00000155205.2
ENSMUST00000120105.8 |
Cdk9
|
cyclin-dependent kinase 9 (CDC2-related kinase) |
| chr11_-_70873773 | 0.11 |
ENSMUST00000078528.7
|
C1qbp
|
complement component 1, q subcomponent binding protein |
| chr8_+_22682816 | 0.11 |
ENSMUST00000033866.9
|
Vps36
|
vacuolar protein sorting 36 |
| chr3_-_84063067 | 0.11 |
ENSMUST00000047368.8
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr13_+_119624575 | 0.10 |
ENSMUST00000176171.9
ENSMUST00000223912.2 |
Tmem267
|
transmembrane protein 267 |
| chr5_-_143301113 | 0.10 |
ENSMUST00000046418.3
|
E130309D02Rik
|
RIKEN cDNA E130309D02 gene |
| chr4_-_149222057 | 0.10 |
ENSMUST00000030813.10
|
Cenps
|
centromere protein S |
| chr14_+_55716227 | 0.10 |
ENSMUST00000227488.2
|
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr10_-_17898938 | 0.10 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr5_+_124045238 | 0.10 |
ENSMUST00000023869.15
|
Denr
|
density-regulated protein |
| chr16_-_10603389 | 0.10 |
ENSMUST00000229866.2
ENSMUST00000038099.6 |
Socs1
|
suppressor of cytokine signaling 1 |
| chrX_-_72502595 | 0.10 |
ENSMUST00000033737.15
ENSMUST00000077243.5 |
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr2_+_93017916 | 0.10 |
ENSMUST00000090554.11
|
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr18_-_10610048 | 0.10 |
ENSMUST00000115864.8
ENSMUST00000145320.2 ENSMUST00000097670.10 |
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr5_-_123270702 | 0.10 |
ENSMUST00000031401.6
|
Rhof
|
ras homolog family member F (in filopodia) |
| chr12_+_24758968 | 0.10 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr15_+_76253084 | 0.10 |
ENSMUST00000023213.8
|
Hgh1
|
HGH1 homolog |
| chr18_+_34910064 | 0.10 |
ENSMUST00000043775.9
ENSMUST00000224715.2 |
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr16_+_16120810 | 0.10 |
ENSMUST00000159962.8
ENSMUST00000059955.15 |
Yars2
|
tyrosyl-tRNA synthetase 2 (mitochondrial) |
| chr9_+_45817795 | 0.10 |
ENSMUST00000039059.8
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr11_-_76468527 | 0.10 |
ENSMUST00000176179.8
|
Abr
|
active BCR-related gene |
| chr8_-_105261262 | 0.10 |
ENSMUST00000162466.8
ENSMUST00000034349.10 |
Nae1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr17_+_27136065 | 0.10 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chrX_-_10083157 | 0.10 |
ENSMUST00000072393.9
ENSMUST00000044598.13 ENSMUST00000073392.11 ENSMUST00000115533.8 ENSMUST00000115532.2 |
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr12_+_24758724 | 0.10 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr7_+_16043502 | 0.10 |
ENSMUST00000002152.13
|
Bbc3
|
BCL2 binding component 3 |
| chr19_+_7394951 | 0.10 |
ENSMUST00000159348.3
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr3_-_97517472 | 0.09 |
ENSMUST00000029730.5
|
Chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr7_-_44785603 | 0.09 |
ENSMUST00000209467.2
|
Gm45713
|
predicted gene 45713 |
| chr3_+_14706781 | 0.09 |
ENSMUST00000029071.9
|
Car13
|
carbonic anhydrase 13 |
| chr11_-_82761954 | 0.09 |
ENSMUST00000108173.10
ENSMUST00000071152.14 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr1_-_36596598 | 0.09 |
ENSMUST00000191642.6
ENSMUST00000193382.6 ENSMUST00000195620.6 |
Sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr5_-_114266228 | 0.09 |
ENSMUST00000053657.13
ENSMUST00000149418.2 ENSMUST00000112279.2 |
Alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr16_-_20245138 | 0.09 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr16_-_10993919 | 0.09 |
ENSMUST00000142389.3
ENSMUST00000138185.9 |
Zc3h7a
|
zinc finger CCCH type containing 7 A |
| chr8_+_40876827 | 0.09 |
ENSMUST00000049389.11
ENSMUST00000128166.8 ENSMUST00000167766.2 |
Zdhhc2
|
zinc finger, DHHC domain containing 2 |
| chr5_+_31079177 | 0.09 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr15_-_102259158 | 0.09 |
ENSMUST00000231061.2
ENSMUST00000041208.9 |
Aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr4_-_132072988 | 0.09 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr6_+_89620956 | 0.09 |
ENSMUST00000000828.14
ENSMUST00000101171.3 |
Txnrd3
|
thioredoxin reductase 3 |
| chr16_+_20536545 | 0.09 |
ENSMUST00000231656.2
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr18_+_37898633 | 0.09 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr14_+_8348779 | 0.09 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr3_+_96552895 | 0.09 |
ENSMUST00000119365.8
ENSMUST00000029744.6 |
Itga10
|
integrin, alpha 10 |
| chr9_-_44179758 | 0.09 |
ENSMUST00000034621.16
ENSMUST00000168499.9 |
Nlrx1
|
NLR family member X1 |
| chr5_+_108048697 | 0.08 |
ENSMUST00000153590.2
|
Rpl5
|
ribosomal protein L5 |
| chr9_-_44179348 | 0.08 |
ENSMUST00000169651.3
|
Nlrx1
|
NLR family member X1 |
| chr12_+_24758240 | 0.08 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr19_-_6947076 | 0.08 |
ENSMUST00000237074.2
|
Plcb3
|
phospholipase C, beta 3 |
| chr11_+_62539479 | 0.08 |
ENSMUST00000062860.5
|
Mmgt2
|
membrane magnesium transporter 2 |
| chr7_-_141017525 | 0.08 |
ENSMUST00000106006.8
ENSMUST00000201710.4 |
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chrX_+_158315666 | 0.08 |
ENSMUST00000057180.13
|
Bclaf3
|
Bclaf1 and Thrap3 family member 3 |
| chr9_+_110162470 | 0.08 |
ENSMUST00000198761.5
ENSMUST00000197630.3 |
Scap
|
SREBF chaperone |
| chr6_+_88061464 | 0.08 |
ENSMUST00000032143.8
|
Rpn1
|
ribophorin I |
| chr2_-_11506893 | 0.08 |
ENSMUST00000114845.10
ENSMUST00000171188.9 ENSMUST00000179584.8 ENSMUST00000028114.13 ENSMUST00000114846.9 ENSMUST00000170196.9 ENSMUST00000191668.6 ENSMUST00000049849.12 ENSMUST00000114844.8 ENSMUST00000100411.4 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr9_+_106088798 | 0.08 |
ENSMUST00000216850.2
|
Twf2
|
twinfilin actin binding protein 2 |
| chr4_-_132073048 | 0.08 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr4_+_41762309 | 0.08 |
ENSMUST00000108042.3
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr7_-_120269462 | 0.08 |
ENSMUST00000127845.2
ENSMUST00000208635.2 ENSMUST00000033178.4 |
Pdzd9
|
PDZ domain containing 9 |
| chr17_-_34109513 | 0.08 |
ENSMUST00000173386.2
ENSMUST00000114361.9 ENSMUST00000173492.9 |
Kifc1
|
kinesin family member C1 |
| chr8_-_25592001 | 0.08 |
ENSMUST00000128715.8
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr2_+_122479770 | 0.08 |
ENSMUST00000047498.15
ENSMUST00000110512.4 |
AA467197
|
expressed sequence AA467197 |
| chr11_+_83363905 | 0.07 |
ENSMUST00000021018.11
|
Taf15
|
TATA-box binding protein associated factor 15 |
| chr4_+_126156118 | 0.07 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr16_+_20536415 | 0.07 |
ENSMUST00000021405.8
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr10_-_88214913 | 0.07 |
ENSMUST00000138159.2
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chrX_+_158315652 | 0.07 |
ENSMUST00000112464.8
|
Bclaf3
|
Bclaf1 and Thrap3 family member 3 |
| chr5_+_115697526 | 0.07 |
ENSMUST00000086519.12
ENSMUST00000156359.2 ENSMUST00000152976.2 |
Rplp0
|
ribosomal protein, large, P0 |
| chr18_+_12776358 | 0.07 |
ENSMUST00000234966.2
ENSMUST00000025294.9 |
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr14_+_55716377 | 0.07 |
ENSMUST00000226902.2
ENSMUST00000228353.2 |
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chrX_-_139501246 | 0.07 |
ENSMUST00000112996.9
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr7_-_16761732 | 0.07 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr17_+_34423054 | 0.07 |
ENSMUST00000138491.2
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr4_+_11704438 | 0.07 |
ENSMUST00000108304.9
|
Gem
|
GTP binding protein (gene overexpressed in skeletal muscle) |
| chr12_-_102671154 | 0.07 |
ENSMUST00000178697.2
ENSMUST00000046518.12 |
Itpk1
|
inositol 1,3,4-triphosphate 5/6 kinase |
| chrX_-_101232978 | 0.07 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr5_+_124045552 | 0.07 |
ENSMUST00000166233.2
|
Denr
|
density-regulated protein |
| chr8_-_105169621 | 0.07 |
ENSMUST00000041769.8
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chrX_+_135950334 | 0.07 |
ENSMUST00000047852.8
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr10_+_53473032 | 0.07 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr3_+_154302311 | 0.07 |
ENSMUST00000192462.6
ENSMUST00000029850.15 |
Cryz
|
crystallin, zeta |
| chr6_+_134988572 | 0.06 |
ENSMUST00000032326.11
ENSMUST00000130851.8 ENSMUST00000205244.3 ENSMUST00000205055.3 ENSMUST00000204646.3 ENSMUST00000154558.3 |
Ddx47
|
DEAD box helicase 47 |
| chr4_+_108317197 | 0.06 |
ENSMUST00000097925.9
|
Tut4
|
terminal uridylyl transferase 4 |
| chr2_-_152673585 | 0.06 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr2_-_4656870 | 0.06 |
ENSMUST00000192470.2
ENSMUST00000035721.14 ENSMUST00000152362.7 |
Prpf18
|
pre-mRNA processing factor 18 |
| chr19_-_6947112 | 0.06 |
ENSMUST00000025912.10
|
Plcb3
|
phospholipase C, beta 3 |
| chr16_-_26190578 | 0.06 |
ENSMUST00000023154.3
|
Cldn1
|
claudin 1 |
| chr5_+_3593811 | 0.06 |
ENSMUST00000197082.5
ENSMUST00000115527.8 |
Fam133b
|
family with sequence similarity 133, member B |
| chr9_-_100428113 | 0.06 |
ENSMUST00000186591.7
ENSMUST00000116522.8 |
Nck1
|
non-catalytic region of tyrosine kinase adaptor protein 1 |
| chr13_+_51254852 | 0.06 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr18_+_73706115 | 0.06 |
ENSMUST00000091852.5
|
Mex3c
|
mex3 RNA binding family member C |
| chr11_-_107080150 | 0.06 |
ENSMUST00000106757.8
ENSMUST00000018577.8 |
Nol11
|
nucleolar protein 11 |
| chr10_+_3923086 | 0.06 |
ENSMUST00000117291.8
ENSMUST00000120585.8 ENSMUST00000043735.8 |
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr14_+_55716302 | 0.06 |
ENSMUST00000226168.2
|
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr19_+_6450553 | 0.06 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr10_+_29189159 | 0.06 |
ENSMUST00000160399.8
|
Echdc1
|
enoyl Coenzyme A hydratase domain containing 1 |
| chr19_+_5012362 | 0.06 |
ENSMUST00000236917.2
|
Mrpl11
|
mitochondrial ribosomal protein L11 |
| chr9_-_43017249 | 0.06 |
ENSMUST00000165665.9
|
Arhgef12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr8_+_34089597 | 0.06 |
ENSMUST00000009774.11
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| chr11_-_100986192 | 0.06 |
ENSMUST00000019447.15
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr11_+_69523754 | 0.06 |
ENSMUST00000018909.4
|
Fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr10_+_53472853 | 0.06 |
ENSMUST00000219271.2
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr7_-_79989965 | 0.06 |
ENSMUST00000127997.2
ENSMUST00000032748.15 |
Unc45a
|
unc-45 myosin chaperone A |
| chr9_-_64160899 | 0.06 |
ENSMUST00000005066.9
|
Map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr3_+_137624231 | 0.06 |
ENSMUST00000197064.5
|
Lamtor3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr14_+_55716197 | 0.06 |
ENSMUST00000022821.8
|
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr7_-_46360066 | 0.06 |
ENSMUST00000143082.4
|
Saal1
|
serum amyloid A-like 1 |
| chr10_+_11157047 | 0.06 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr2_+_28403255 | 0.06 |
ENSMUST00000028170.15
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr6_-_71376277 | 0.06 |
ENSMUST00000149415.2
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr5_+_123390149 | 0.06 |
ENSMUST00000121964.8
|
Wdr66
|
WD repeat domain 66 |
| chr9_-_109930078 | 0.06 |
ENSMUST00000196455.2
ENSMUST00000062368.13 |
Dhx30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30 |
| chr8_+_70735477 | 0.06 |
ENSMUST00000087467.12
ENSMUST00000140212.8 ENSMUST00000110124.9 |
Homer3
|
homer scaffolding protein 3 |
| chr11_-_69470139 | 0.05 |
ENSMUST00000048139.12
|
Wrap53
|
WD repeat containing, antisense to Trp53 |
| chr19_-_10282218 | 0.05 |
ENSMUST00000039327.11
|
Dagla
|
diacylglycerol lipase, alpha |
| chr1_+_191450507 | 0.05 |
ENSMUST00000130876.2
|
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr15_-_89080643 | 0.05 |
ENSMUST00000078953.9
|
Dennd6b
|
DENN/MADD domain containing 6B |
| chr1_-_16590244 | 0.05 |
ENSMUST00000144138.4
ENSMUST00000145092.8 ENSMUST00000131257.9 ENSMUST00000153966.9 ENSMUST00000162435.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr1_+_161796854 | 0.05 |
ENSMUST00000160881.2
ENSMUST00000159648.2 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr4_-_123644091 | 0.05 |
ENSMUST00000102636.4
|
Akirin1
|
akirin 1 |
| chr6_+_29853745 | 0.05 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr10_-_17898838 | 0.05 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr6_-_52217821 | 0.05 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr5_-_65090893 | 0.05 |
ENSMUST00000197315.5
|
Tlr1
|
toll-like receptor 1 |
| chr4_-_45108038 | 0.05 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr14_-_70405288 | 0.05 |
ENSMUST00000129174.8
ENSMUST00000125300.3 |
Pdlim2
|
PDZ and LIM domain 2 |
| chr12_+_76812301 | 0.05 |
ENSMUST00000041262.14
ENSMUST00000126408.2 ENSMUST00000110399.3 ENSMUST00000137826.8 |
Churc1
Fntb
|
churchill domain containing 1 farnesyltransferase, CAAX box, beta |
| chr3_+_142266113 | 0.05 |
ENSMUST00000106221.8
|
Gbp3
|
guanylate binding protein 3 |
| chr14_-_24537067 | 0.05 |
ENSMUST00000026322.9
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr11_+_115705550 | 0.05 |
ENSMUST00000021134.10
ENSMUST00000106481.9 |
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chr3_+_81839908 | 0.05 |
ENSMUST00000029649.3
|
Ctso
|
cathepsin O |
| chr16_+_32698149 | 0.05 |
ENSMUST00000023489.11
ENSMUST00000171325.9 |
Fyttd1
|
forty-two-three domain containing 1 |
| chr15_-_89258012 | 0.05 |
ENSMUST00000167643.4
|
Sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr5_+_140317615 | 0.05 |
ENSMUST00000071881.10
ENSMUST00000050205.12 ENSMUST00000110827.8 |
Nudt1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr10_+_4296806 | 0.05 |
ENSMUST00000216139.3
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr1_+_191449946 | 0.05 |
ENSMUST00000133076.7
ENSMUST00000110855.8 |
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr5_+_105879620 | 0.05 |
ENSMUST00000127686.8
|
Lrrc8d
|
leucine rich repeat containing 8D |
| chr11_-_82799186 | 0.05 |
ENSMUST00000103213.10
|
Nle1
|
notchless homolog 1 |
| chr17_-_56343625 | 0.05 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ctcfl_Ctcf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 0.3 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.2 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.2 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.2 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0002481 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) cytosol to ER transport(GO:0046967) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) |
| 0.0 | 0.1 | GO:0098963 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.0 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0099541 | diacylglycerol catabolic process(GO:0046340) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.0 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.4 | GO:0030677 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.1 | 0.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.1 | 0.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.3 | GO:0018455 | alcohol dehydrogenase [NAD(P)+] activity(GO:0018455) |
| 0.0 | 0.3 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.1 | GO:0051747 | cytosine C-5 DNA demethylase activity(GO:0051747) |
| 0.0 | 0.2 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.0 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |