Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Cux1
Z-value: 0.45
Transcription factors associated with Cux1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cux1
|
ENSMUSG00000029705.18 | cut-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cux1 | mm39_v1_chr5_-_136596094_136596141 | 0.66 | 2.3e-01 | Click! |
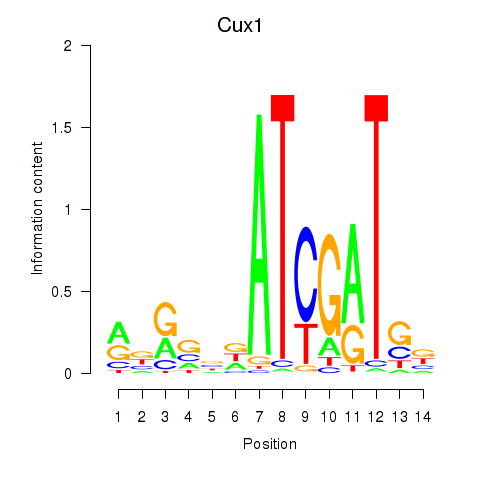
Activity profile of Cux1 motif
Sorted Z-values of Cux1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_21901791 | 0.42 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr13_+_23765546 | 0.41 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4 |
| chr7_+_43000765 | 0.37 |
ENSMUST00000012798.14
ENSMUST00000122423.8 |
Siglecf
|
sialic acid binding Ig-like lectin F |
| chr7_+_43000838 | 0.32 |
ENSMUST00000206299.2
ENSMUST00000121494.2 |
Siglecf
|
sialic acid binding Ig-like lectin F |
| chr13_-_21900313 | 0.30 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr13_+_21994588 | 0.26 |
ENSMUST00000091745.6
|
H2ac23
|
H2A clustered histone 23 |
| chr7_-_3551003 | 0.22 |
ENSMUST00000065703.9
ENSMUST00000203020.3 ENSMUST00000203821.3 |
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chrM_+_9870 | 0.20 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr9_-_108911438 | 0.17 |
ENSMUST00000192801.2
|
Tma7
|
translational machinery associated 7 |
| chr9_+_118892497 | 0.17 |
ENSMUST00000141185.8
ENSMUST00000126251.8 ENSMUST00000136561.2 |
Vill
|
villin-like |
| chr17_+_24955613 | 0.17 |
ENSMUST00000115262.9
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chrM_+_10167 | 0.16 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrM_+_7006 | 0.15 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr8_+_69333143 | 0.15 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase |
| chr13_-_46118433 | 0.15 |
ENSMUST00000167708.4
ENSMUST00000091628.11 ENSMUST00000180110.9 |
Atxn1
|
ataxin 1 |
| chr2_-_18042548 | 0.15 |
ENSMUST00000066163.3
|
A930004D18Rik
|
RIKEN cDNA A930004D18 gene |
| chr7_-_19432308 | 0.14 |
ENSMUST00000173739.8
|
Apoe
|
apolipoprotein E |
| chr2_-_165997179 | 0.14 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr2_-_165996716 | 0.13 |
ENSMUST00000139266.2
|
Sulf2
|
sulfatase 2 |
| chr2_-_165997551 | 0.13 |
ENSMUST00000109249.9
ENSMUST00000146497.9 |
Sulf2
|
sulfatase 2 |
| chr17_+_24955647 | 0.12 |
ENSMUST00000101800.7
|
Msrb1
|
methionine sulfoxide reductase B1 |
| chrM_+_3906 | 0.12 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr17_+_31329939 | 0.11 |
ENSMUST00000236241.2
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr1_+_16758629 | 0.11 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr8_-_32440071 | 0.11 |
ENSMUST00000207678.3
|
Nrg1
|
neuregulin 1 |
| chrM_+_7779 | 0.11 |
ENSMUST00000082408.1
|
mt-Atp6
|
mitochondrially encoded ATP synthase 6 |
| chr4_-_138053603 | 0.10 |
ENSMUST00000030536.13
|
Pink1
|
PTEN induced putative kinase 1 |
| chr16_+_44586085 | 0.10 |
ENSMUST00000057488.15
|
Cd200r1
|
CD200 receptor 1 |
| chr11_-_3881995 | 0.10 |
ENSMUST00000020710.11
ENSMUST00000109989.10 ENSMUST00000109991.8 ENSMUST00000109993.9 |
Tcn2
|
transcobalamin 2 |
| chr1_-_135615831 | 0.10 |
ENSMUST00000190298.8
|
Nav1
|
neuron navigator 1 |
| chr7_+_86645323 | 0.09 |
ENSMUST00000233714.2
ENSMUST00000233648.2 ENSMUST00000164462.3 ENSMUST00000233730.2 |
Vmn2r79
|
vomeronasal 2, receptor 79 |
| chr7_-_3828640 | 0.09 |
ENSMUST00000189095.7
ENSMUST00000094911.5 ENSMUST00000153846.8 ENSMUST00000108619.8 ENSMUST00000108620.8 |
Gm15448
|
predicted gene 15448 |
| chr6_-_57992144 | 0.09 |
ENSMUST00000228070.2
ENSMUST00000228040.2 |
Vmn1r26
|
vomeronasal 1 receptor 26 |
| chr7_-_26878260 | 0.09 |
ENSMUST00000093040.13
|
Rab4b
|
RAB4B, member RAS oncogene family |
| chr17_-_24005020 | 0.09 |
ENSMUST00000024704.10
|
Flywch2
|
FLYWCH family member 2 |
| chr19_+_5651882 | 0.09 |
ENSMUST00000025864.11
ENSMUST00000236461.2 ENSMUST00000155639.2 |
Rnaseh2c
|
ribonuclease H2, subunit C |
| chrM_+_11735 | 0.08 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr15_-_35938328 | 0.08 |
ENSMUST00000014457.15
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chrX_+_150784841 | 0.08 |
ENSMUST00000026289.10
ENSMUST00000112617.4 |
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr3_-_58433313 | 0.08 |
ENSMUST00000029385.9
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr1_-_78488795 | 0.08 |
ENSMUST00000170511.3
|
BC035947
|
cDNA sequence BC035947 |
| chr4_-_138053545 | 0.08 |
ENSMUST00000105817.4
|
Pink1
|
PTEN induced putative kinase 1 |
| chr9_-_58648826 | 0.08 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein |
| chr1_+_171668173 | 0.08 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr11_-_120604551 | 0.07 |
ENSMUST00000106154.8
ENSMUST00000106155.4 ENSMUST00000055424.13 ENSMUST00000026137.8 |
Cenpx
|
centromere protein X |
| chr5_+_144192033 | 0.07 |
ENSMUST00000056578.7
|
Bri3
|
brain protein I3 |
| chr7_+_45683122 | 0.07 |
ENSMUST00000033121.7
|
Nomo1
|
nodal modulator 1 |
| chrM_+_7758 | 0.07 |
ENSMUST00000082407.1
|
mt-Atp8
|
mitochondrially encoded ATP synthase 8 |
| chr2_-_76503382 | 0.07 |
ENSMUST00000002809.14
|
Fkbp7
|
FK506 binding protein 7 |
| chr6_-_88021999 | 0.07 |
ENSMUST00000113598.8
|
Rab7
|
RAB7, member RAS oncogene family |
| chr11_-_16458484 | 0.07 |
ENSMUST00000109643.8
ENSMUST00000166950.8 ENSMUST00000178855.8 |
Sec61g
|
SEC61, gamma subunit |
| chr11_-_109502243 | 0.07 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chrX_-_73436814 | 0.07 |
ENSMUST00000122894.8
ENSMUST00000114143.10 |
Fam3a
|
family with sequence similarity 3, member A |
| chr12_-_114451189 | 0.06 |
ENSMUST00000103493.3
|
Ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr1_+_16758731 | 0.06 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chrX_-_105264751 | 0.06 |
ENSMUST00000113495.9
|
Taf9b
|
TATA-box binding protein associated factor 9B |
| chr6_-_118116062 | 0.06 |
ENSMUST00000049344.15
|
Csgalnact2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr10_-_117118226 | 0.06 |
ENSMUST00000092163.9
|
Lyz2
|
lysozyme 2 |
| chr11_-_76969230 | 0.06 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1 |
| chr12_-_112905721 | 0.06 |
ENSMUST00000221497.2
ENSMUST00000002881.5 ENSMUST00000221397.2 |
Nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr1_-_179373826 | 0.06 |
ENSMUST00000027769.6
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr2_+_90927053 | 0.06 |
ENSMUST00000132741.3
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr5_-_3646071 | 0.06 |
ENSMUST00000121877.3
|
Rbm48
|
RNA binding motif protein 48 |
| chr18_+_39439778 | 0.06 |
ENSMUST00000235660.2
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr5_-_3646540 | 0.06 |
ENSMUST00000042753.14
|
Rbm48
|
RNA binding motif protein 48 |
| chr7_+_44033520 | 0.06 |
ENSMUST00000118962.8
ENSMUST00000118831.8 |
Syt3
|
synaptotagmin III |
| chr10_+_79715448 | 0.06 |
ENSMUST00000006679.15
|
Prtn3
|
proteinase 3 |
| chr7_-_41708447 | 0.06 |
ENSMUST00000168489.3
ENSMUST00000233456.2 |
Vmn2r59
|
vomeronasal 2, receptor 59 |
| chr2_+_178056302 | 0.06 |
ENSMUST00000094251.11
|
Fam217b
|
family with sequence similarity 217, member B |
| chr2_-_178056251 | 0.05 |
ENSMUST00000058678.5
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr10_-_126956991 | 0.05 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chrX_+_106192510 | 0.05 |
ENSMUST00000147521.8
ENSMUST00000167673.2 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr5_-_130031681 | 0.05 |
ENSMUST00000111308.8
ENSMUST00000111307.2 |
Gusb
|
glucuronidase, beta |
| chr3_+_144283355 | 0.05 |
ENSMUST00000151086.3
|
Selenof
|
selenoprotein F |
| chr19_+_44282113 | 0.05 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr3_+_28835425 | 0.05 |
ENSMUST00000060500.9
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr17_+_46991906 | 0.05 |
ENSMUST00000233430.2
|
Mea1
|
male enhanced antigen 1 |
| chr15_-_89239858 | 0.05 |
ENSMUST00000023283.6
|
Lmf2
|
lipase maturation factor 2 |
| chr1_+_157353696 | 0.05 |
ENSMUST00000111700.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr2_+_151947444 | 0.05 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chrX_+_7765589 | 0.05 |
ENSMUST00000033498.10
|
Timm17b
|
translocase of inner mitochondrial membrane 17b |
| chr7_+_116103643 | 0.05 |
ENSMUST00000183175.8
|
Nucb2
|
nucleobindin 2 |
| chrX_+_105059305 | 0.05 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr7_-_110681402 | 0.05 |
ENSMUST00000159305.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr4_-_155953849 | 0.05 |
ENSMUST00000030950.8
|
Cptp
|
ceramide-1-phosphate transfer protein |
| chr3_-_88204145 | 0.05 |
ENSMUST00000010682.4
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr2_+_132528851 | 0.05 |
ENSMUST00000061891.11
|
Shld1
|
shieldin complex subunit 1 |
| chr7_+_116103599 | 0.05 |
ENSMUST00000032895.15
|
Nucb2
|
nucleobindin 2 |
| chr19_-_4889284 | 0.05 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr15_+_43340609 | 0.05 |
ENSMUST00000022962.8
|
Emc2
|
ER membrane protein complex subunit 2 |
| chr5_+_54155814 | 0.05 |
ENSMUST00000117661.9
|
Stim2
|
stromal interaction molecule 2 |
| chrX_+_106193167 | 0.05 |
ENSMUST00000137107.2
ENSMUST00000067249.3 |
P2ry10b
|
purinergic receptor P2Y, G-protein coupled 10B |
| chr11_-_95966477 | 0.05 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr9_-_44831995 | 0.05 |
ENSMUST00000043675.9
|
Atp5l
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit G |
| chr4_-_117729717 | 0.05 |
ENSMUST00000171052.8
ENSMUST00000166325.2 ENSMUST00000106422.9 |
Ccdc24
|
coiled-coil domain containing 24 |
| chr3_+_89173859 | 0.05 |
ENSMUST00000040824.2
|
Dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr16_-_35311243 | 0.05 |
ENSMUST00000023550.9
|
Pdia5
|
protein disulfide isomerase associated 5 |
| chr17_-_22152003 | 0.04 |
ENSMUST00000074295.8
|
Zfp942
|
zinc finger protein 942 |
| chr9_+_59446823 | 0.04 |
ENSMUST00000026262.8
|
Hexa
|
hexosaminidase A |
| chr1_-_169358912 | 0.04 |
ENSMUST00000192248.2
ENSMUST00000028000.13 |
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr13_-_74882374 | 0.04 |
ENSMUST00000220738.2
|
Cast
|
calpastatin |
| chr13_-_43324937 | 0.04 |
ENSMUST00000222160.2
ENSMUST00000179852.9 ENSMUST00000021797.9 |
Tbc1d7
|
TBC1 domain family, member 7 |
| chr17_+_46991972 | 0.04 |
ENSMUST00000002845.8
|
Mea1
|
male enhanced antigen 1 |
| chr6_-_67468831 | 0.04 |
ENSMUST00000118364.2
|
Il23r
|
interleukin 23 receptor |
| chr17_-_24388384 | 0.04 |
ENSMUST00000024932.12
|
Atp6v0c
|
ATPase, H+ transporting, lysosomal V0 subunit C |
| chr7_-_6525801 | 0.04 |
ENSMUST00000213504.2
ENSMUST00000216447.2 ENSMUST00000213656.2 ENSMUST00000207820.3 |
Olfr1349
|
olfactory receptor 1349 |
| chr3_-_143910926 | 0.04 |
ENSMUST00000120539.8
ENSMUST00000196264.5 |
Lmo4
|
LIM domain only 4 |
| chr1_-_182345011 | 0.04 |
ENSMUST00000068505.10
|
Capn2
|
calpain 2 |
| chr3_-_103698391 | 0.04 |
ENSMUST00000106845.9
ENSMUST00000029438.15 |
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr10_+_80003612 | 0.04 |
ENSMUST00000105365.9
|
Cirbp
|
cold inducible RNA binding protein |
| chr14_+_118374511 | 0.04 |
ENSMUST00000022728.4
|
Gpr180
|
G protein-coupled receptor 180 |
| chr4_+_63462984 | 0.04 |
ENSMUST00000035301.7
|
Atp6v1g1
|
ATPase, H+ transporting, lysosomal V1 subunit G1 |
| chrM_+_8603 | 0.04 |
ENSMUST00000082409.1
|
mt-Co3
|
mitochondrially encoded cytochrome c oxidase III |
| chrX_-_69520778 | 0.04 |
ENSMUST00000101506.10
ENSMUST00000114630.3 |
Tmem185a
|
transmembrane protein 185A |
| chr5_+_109567554 | 0.04 |
ENSMUST00000232833.2
ENSMUST00000232722.2 ENSMUST00000233536.2 ENSMUST00000171841.3 ENSMUST00000233038.2 ENSMUST00000233724.2 |
Vmn2r17
|
vomeronasal 2, receptor 17 |
| chr14_+_73411249 | 0.04 |
ENSMUST00000166875.2
|
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr14_+_70314727 | 0.04 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr6_+_124908389 | 0.04 |
ENSMUST00000180095.4
|
Mlf2
|
myeloid leukemia factor 2 |
| chr8_+_71849497 | 0.04 |
ENSMUST00000002473.10
|
Babam1
|
BRISC and BRCA1 A complex member 1 |
| chr8_-_41827639 | 0.04 |
ENSMUST00000034000.15
ENSMUST00000143057.2 |
Asah1
|
N-acylsphingosine amidohydrolase 1 |
| chr5_-_122188165 | 0.04 |
ENSMUST00000154139.8
|
Cux2
|
cut-like homeobox 2 |
| chr13_+_74554509 | 0.04 |
ENSMUST00000222435.2
|
Ftl1-ps1
|
ferritin light polypeptide 1, pseudogene 1 |
| chr13_-_74882328 | 0.04 |
ENSMUST00000223309.2
|
Cast
|
calpastatin |
| chr5_+_54155871 | 0.04 |
ENSMUST00000201469.4
|
Stim2
|
stromal interaction molecule 2 |
| chr11_+_78389913 | 0.04 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin |
| chr8_-_78337226 | 0.04 |
ENSMUST00000034030.15
|
Tmem184c
|
transmembrane protein 184C |
| chr19_-_7183596 | 0.04 |
ENSMUST00000123594.8
|
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr3_-_88204286 | 0.04 |
ENSMUST00000107556.10
|
Tsacc
|
TSSK6 activating co-chaperone |
| chr10_-_127358231 | 0.04 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr10_+_85707687 | 0.04 |
ENSMUST00000001836.11
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr2_-_25465236 | 0.04 |
ENSMUST00000039156.7
|
Phpt1
|
phosphohistidine phosphatase 1 |
| chr11_-_62348115 | 0.04 |
ENSMUST00000069456.11
ENSMUST00000018645.13 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr6_+_124908439 | 0.04 |
ENSMUST00000032214.14
|
Mlf2
|
myeloid leukemia factor 2 |
| chr12_-_115109539 | 0.04 |
ENSMUST00000192554.6
ENSMUST00000103522.3 |
Ighv1-52
|
immunoglobulin heavy variable 1-52 |
| chr10_+_88036947 | 0.04 |
ENSMUST00000020248.16
ENSMUST00000182183.8 ENSMUST00000171151.9 ENSMUST00000182619.2 |
Washc3
|
WASH complex subunit 3 |
| chr6_+_124908341 | 0.04 |
ENSMUST00000203021.3
|
Mlf2
|
myeloid leukemia factor 2 |
| chr10_-_127358300 | 0.04 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr8_-_71060911 | 0.04 |
ENSMUST00000210580.2
ENSMUST00000211608.2 ENSMUST00000049908.11 |
Ssbp4
|
single stranded DNA binding protein 4 |
| chr9_+_108765701 | 0.04 |
ENSMUST00000026743.14
ENSMUST00000194047.3 |
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein 1 |
| chr3_-_89121686 | 0.04 |
ENSMUST00000073572.12
|
Mtx1
|
metaxin 1 |
| chr19_-_13126896 | 0.04 |
ENSMUST00000213493.2
|
Olfr1459
|
olfactory receptor 1459 |
| chr7_+_140972070 | 0.03 |
ENSMUST00000211654.2
ENSMUST00000026576.5 |
Taldo1
|
transaldolase 1 |
| chr10_+_80165787 | 0.03 |
ENSMUST00000105358.8
ENSMUST00000105357.2 ENSMUST00000105354.8 ENSMUST00000105355.8 |
Reep6
|
receptor accessory protein 6 |
| chr10_+_80084955 | 0.03 |
ENSMUST00000105364.8
|
Ndufs7
|
NADH:ubiquinone oxidoreductase core subunit S7 |
| chr5_-_108017019 | 0.03 |
ENSMUST00000128723.8
ENSMUST00000124034.8 |
Evi5
|
ecotropic viral integration site 5 |
| chr2_-_69715899 | 0.03 |
ENSMUST00000060447.13
|
Mettl5
|
methyltransferase like 5 |
| chrX_-_73436786 | 0.03 |
ENSMUST00000114141.8
|
Fam3a
|
family with sequence similarity 3, member A |
| chr2_+_174169539 | 0.03 |
ENSMUST00000133356.8
ENSMUST00000087871.11 |
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus |
| chr2_-_168797430 | 0.03 |
ENSMUST00000109162.9
|
Zfp64
|
zinc finger protein 64 |
| chr3_-_89121637 | 0.03 |
ENSMUST00000239452.2
ENSMUST00000118964.10 |
Mtx1
|
metaxin 1 |
| chr7_+_142606476 | 0.03 |
ENSMUST00000037941.10
|
Cd81
|
CD81 antigen |
| chr9_+_120400510 | 0.03 |
ENSMUST00000165532.3
|
Rpl14
|
ribosomal protein L14 |
| chr2_+_65676111 | 0.03 |
ENSMUST00000122912.8
|
Csrnp3
|
cysteine-serine-rich nuclear protein 3 |
| chr18_+_70701260 | 0.03 |
ENSMUST00000074058.11
ENSMUST00000114946.4 |
Mbd2
|
methyl-CpG binding domain protein 2 |
| chr5_-_32903687 | 0.03 |
ENSMUST00000135248.2
|
Pisd
|
phosphatidylserine decarboxylase |
| chr11_-_106378659 | 0.03 |
ENSMUST00000106801.8
|
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1 |
| chr18_+_67523734 | 0.03 |
ENSMUST00000001513.8
|
Tubb6
|
tubulin, beta 6 class V |
| chr6_+_149310471 | 0.03 |
ENSMUST00000086829.11
ENSMUST00000111513.9 |
Bicd1
|
BICD cargo adaptor 1 |
| chr13_+_108452930 | 0.03 |
ENSMUST00000171178.2
|
Depdc1b
|
DEP domain containing 1B |
| chr14_+_101891416 | 0.03 |
ENSMUST00000002289.8
|
Uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr15_-_36609812 | 0.03 |
ENSMUST00000226496.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr9_-_41068771 | 0.03 |
ENSMUST00000136530.8
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B |
| chrX_-_93256291 | 0.03 |
ENSMUST00000050328.15
|
Eif2s3x
|
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
| chr11_+_101623776 | 0.03 |
ENSMUST00000039152.14
|
Dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr3_+_132797588 | 0.03 |
ENSMUST00000029650.9
|
Ints12
|
integrator complex subunit 12 |
| chr8_-_110305672 | 0.03 |
ENSMUST00000074898.8
|
Hp
|
haptoglobin |
| chr13_-_35211060 | 0.03 |
ENSMUST00000170538.8
ENSMUST00000163280.8 |
Eci2
|
enoyl-Coenzyme A delta isomerase 2 |
| chr17_+_34866160 | 0.03 |
ENSMUST00000173984.2
|
Atf6b
|
activating transcription factor 6 beta |
| chr7_+_4240697 | 0.03 |
ENSMUST00000117550.2
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr10_+_92996686 | 0.03 |
ENSMUST00000069965.9
|
Cdk17
|
cyclin-dependent kinase 17 |
| chr1_+_88128323 | 0.03 |
ENSMUST00000049289.9
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
| chr11_-_106378622 | 0.03 |
ENSMUST00000001059.9
ENSMUST00000106799.2 ENSMUST00000106800.2 |
Ern1
|
endoplasmic reticulum (ER) to nucleus signalling 1 |
| chr8_-_71349927 | 0.03 |
ENSMUST00000212709.2
ENSMUST00000212796.2 ENSMUST00000212378.2 ENSMUST00000054220.10 ENSMUST00000212494.2 |
Rpl18a
|
ribosomal protein L18A |
| chr8_+_61677253 | 0.03 |
ENSMUST00000209611.2
|
Sh3rf1
|
SH3 domain containing ring finger 1 |
| chrX_-_167456828 | 0.03 |
ENSMUST00000033725.16
ENSMUST00000112137.2 |
Msl3
|
MSL complex subunit 3 |
| chr9_+_26910995 | 0.03 |
ENSMUST00000213770.2
ENSMUST00000213683.2 ENSMUST00000039161.10 |
Thyn1
|
thymocyte nuclear protein 1 |
| chr1_+_75412574 | 0.03 |
ENSMUST00000037796.14
ENSMUST00000113584.8 ENSMUST00000145166.8 ENSMUST00000143730.8 ENSMUST00000133418.8 ENSMUST00000144874.8 ENSMUST00000140287.8 |
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr8_+_66070661 | 0.03 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr10_-_126866658 | 0.03 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr17_+_34866090 | 0.03 |
ENSMUST00000015605.15
|
Atf6b
|
activating transcription factor 6 beta |
| chr11_-_83189670 | 0.03 |
ENSMUST00000175741.8
ENSMUST00000176518.2 |
Pex12
|
peroxisomal biogenesis factor 12 |
| chr9_-_57672375 | 0.03 |
ENSMUST00000215233.2
|
Clk3
|
CDC-like kinase 3 |
| chr2_-_118310116 | 0.03 |
ENSMUST00000110862.2
ENSMUST00000009693.15 |
Srp14
|
signal recognition particle 14 |
| chr9_-_100916910 | 0.03 |
ENSMUST00000142676.8
ENSMUST00000149322.8 ENSMUST00000189498.2 |
Pccb
|
propionyl Coenzyme A carboxylase, beta polypeptide |
| chr15_-_36598263 | 0.03 |
ENSMUST00000155116.2
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr5_+_122845607 | 0.03 |
ENSMUST00000031429.14
|
P2rx4
|
purinergic receptor P2X, ligand-gated ion channel 4 |
| chr1_-_119765068 | 0.03 |
ENSMUST00000163435.8
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr6_+_132844971 | 0.03 |
ENSMUST00000082014.2
|
Tas2r110
|
taste receptor, type 2, member 110 |
| chr4_+_155915729 | 0.03 |
ENSMUST00000139651.8
ENSMUST00000084097.12 |
Aurkaip1
|
aurora kinase A interacting protein 1 |
| chr19_-_4889314 | 0.03 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr1_-_53002409 | 0.03 |
ENSMUST00000114493.2
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr7_-_27373939 | 0.03 |
ENSMUST00000138243.2
|
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr2_-_127630769 | 0.03 |
ENSMUST00000028857.14
ENSMUST00000110357.2 |
Nphp1
|
nephronophthisis 1 (juvenile) homolog (human) |
| chr5_+_8106527 | 0.03 |
ENSMUST00000148633.4
|
Sri
|
sorcin |
| chrX_+_141011173 | 0.03 |
ENSMUST00000112914.8
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr7_-_27374017 | 0.03 |
ENSMUST00000036453.14
ENSMUST00000108341.2 |
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr14_+_105496147 | 0.03 |
ENSMUST00000138283.2
|
Ndfip2
|
Nedd4 family interacting protein 2 |
| chr1_-_34478753 | 0.03 |
ENSMUST00000042493.10
|
Ccdc115
|
coiled-coil domain containing 115 |
| chr17_-_20701593 | 0.03 |
ENSMUST00000167314.3
ENSMUST00000232850.2 ENSMUST00000233382.2 ENSMUST00000233572.2 ENSMUST00000233830.2 |
Vmn2r108
|
vomeronasal 2, receptor 108 |
| chrX_+_47783881 | 0.03 |
ENSMUST00000033433.3
|
Rbmx2
|
RNA binding motif protein, X-linked 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cux1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.0 | 0.1 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.1 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.0 | 0.2 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of macromitophagy(GO:1901526) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:1905034 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.2 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.0 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:1990630 | IRE1-RACK1-PP2A complex(GO:1990630) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.0 | 0.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |