Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Cxxc1
Z-value: 0.38
Transcription factors associated with Cxxc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Cxxc1
|
ENSMUSG00000024560.8 | CXXC finger 1 (PHD domain) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cxxc1 | mm39_v1_chr18_+_74349189_74349217 | -0.71 | 1.8e-01 | Click! |
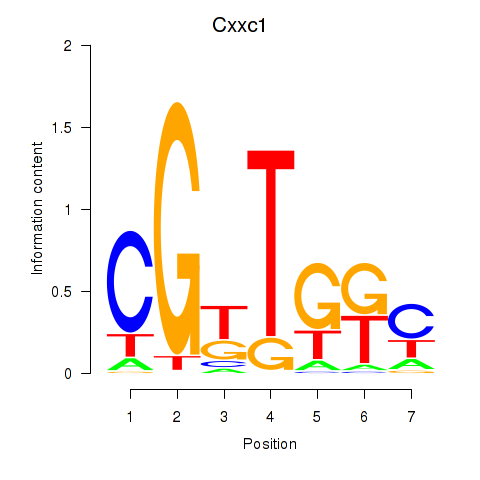
Activity profile of Cxxc1 motif
Sorted Z-values of Cxxc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_96175970 | 0.86 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chr1_+_59296065 | 0.27 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr5_-_25305621 | 0.26 |
ENSMUST00000030784.14
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr5_-_110957909 | 0.16 |
ENSMUST00000200299.2
|
Ulk1
|
unc-51 like kinase 1 |
| chr7_-_45546081 | 0.15 |
ENSMUST00000120299.4
ENSMUST00000039049.16 |
Syngr4
|
synaptogyrin 4 |
| chr7_-_28135616 | 0.10 |
ENSMUST00000208199.2
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr9_+_78016468 | 0.10 |
ENSMUST00000118869.8
ENSMUST00000125615.2 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chr11_+_113539995 | 0.09 |
ENSMUST00000018805.15
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr3_-_146205429 | 0.09 |
ENSMUST00000029839.11
|
Spata1
|
spermatogenesis associated 1 |
| chr5_-_110957933 | 0.08 |
ENSMUST00000031490.11
|
Ulk1
|
unc-51 like kinase 1 |
| chr16_-_64591509 | 0.08 |
ENSMUST00000076991.7
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene |
| chr1_-_5987617 | 0.07 |
ENSMUST00000044180.5
|
Npbwr1
|
neuropeptides B/W receptor 1 |
| chr19_-_19088543 | 0.06 |
ENSMUST00000112832.8
|
Rorb
|
RAR-related orphan receptor beta |
| chr11_+_113540154 | 0.04 |
ENSMUST00000063776.14
|
Cog1
|
component of oligomeric golgi complex 1 |
| chr2_+_151753119 | 0.03 |
ENSMUST00000028955.6
|
Angpt4
|
angiopoietin 4 |
| chr17_-_87754282 | 0.03 |
ENSMUST00000234406.2
ENSMUST00000040440.7 |
Calm2
|
calmodulin 2 |
| chr19_-_36896999 | 0.03 |
ENSMUST00000238948.2
ENSMUST00000057337.9 |
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr4_+_108857803 | 0.03 |
ENSMUST00000065977.11
ENSMUST00000102736.9 |
Nrd1
|
nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
| chr19_+_38919477 | 0.03 |
ENSMUST00000145051.2
|
Hells
|
helicase, lymphoid specific |
| chr11_-_88608958 | 0.02 |
ENSMUST00000107908.2
|
Msi2
|
musashi RNA-binding protein 2 |
| chr5_+_25721059 | 0.02 |
ENSMUST00000045016.9
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr10_+_75790348 | 0.02 |
ENSMUST00000099577.4
|
Gm5134
|
predicted gene 5134 |
| chrX_+_5959507 | 0.02 |
ENSMUST00000103007.4
|
Nudt11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr3_-_41696906 | 0.01 |
ENSMUST00000026866.15
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr1_+_86354045 | 0.01 |
ENSMUST00000046004.6
|
Tex44
|
testis expressed 44 |
| chr7_+_55700089 | 0.01 |
ENSMUST00000164095.3
|
Herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr7_+_128346655 | 0.01 |
ENSMUST00000042942.10
|
Sec23ip
|
Sec23 interacting protein |
| chr10_-_51507527 | 0.00 |
ENSMUST00000219286.2
ENSMUST00000020062.4 ENSMUST00000218684.2 |
Gprc6a
|
G protein-coupled receptor, family C, group 6, member A |
| chr7_-_129867967 | 0.00 |
ENSMUST00000117872.8
ENSMUST00000120187.9 |
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr11_-_88609048 | 0.00 |
ENSMUST00000107909.8
|
Msi2
|
musashi RNA-binding protein 2 |
| chr7_-_129867921 | 0.00 |
ENSMUST00000122054.8
|
Fgfr2
|
fibroblast growth factor receptor 2 |
| chr9_-_107483855 | 0.00 |
ENSMUST00000073448.12
ENSMUST00000194606.2 ENSMUST00000195662.6 |
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr14_-_62718581 | 0.00 |
ENSMUST00000186010.8
|
Gm4131
|
predicted gene 4131 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Cxxc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097635 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) extrinsic component of autophagosome membrane(GO:0097635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |