Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
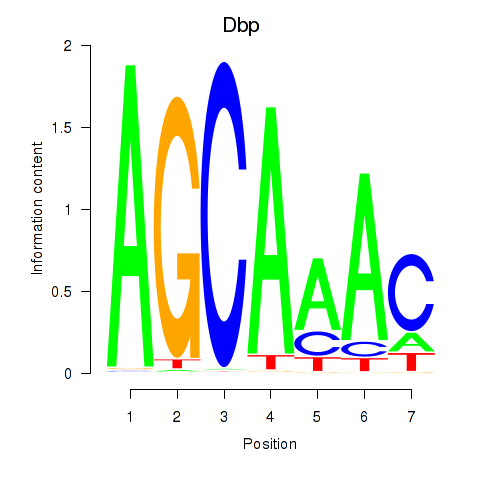
Results for Dbp
Z-value: 0.78
Transcription factors associated with Dbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dbp
|
ENSMUSG00000059824.13 | D site albumin promoter binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dbp | mm39_v1_chr7_+_45354512_45354735 | 0.55 | 3.4e-01 | Click! |
Activity profile of Dbp motif
Sorted Z-values of Dbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_77262968 | 0.75 |
ENSMUST00000081964.7
|
Hopx
|
HOP homeobox |
| chr4_-_87724533 | 0.63 |
ENSMUST00000126353.8
ENSMUST00000149357.8 |
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chrX_+_168468186 | 0.53 |
ENSMUST00000112107.8
ENSMUST00000112104.8 |
Mid1
|
midline 1 |
| chr17_+_26882171 | 0.45 |
ENSMUST00000236346.2
|
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr13_-_29137673 | 0.44 |
ENSMUST00000067230.6
|
Sox4
|
SRY (sex determining region Y)-box 4 |
| chr10_+_99099084 | 0.44 |
ENSMUST00000020118.5
ENSMUST00000220291.2 |
Dusp6
|
dual specificity phosphatase 6 |
| chr7_-_84328553 | 0.44 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr8_-_79539838 | 0.42 |
ENSMUST00000146824.2
|
Lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_+_28813125 | 0.42 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr4_-_87724512 | 0.42 |
ENSMUST00000148059.2
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr13_+_28441511 | 0.41 |
ENSMUST00000223428.2
|
Rps18-ps5
|
ribosomal protein S18, pseudogene 5 |
| chr14_+_79753055 | 0.40 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr4_+_28813152 | 0.39 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chr6_+_145091196 | 0.38 |
ENSMUST00000156849.8
ENSMUST00000132948.2 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr3_-_50398027 | 0.36 |
ENSMUST00000029297.6
ENSMUST00000194462.6 |
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr13_+_94954202 | 0.32 |
ENSMUST00000220825.2
|
Tbca
|
tubulin cofactor A |
| chr11_-_96714813 | 0.32 |
ENSMUST00000142065.2
ENSMUST00000167110.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr18_+_11766333 | 0.31 |
ENSMUST00000115861.9
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr14_+_74973081 | 0.30 |
ENSMUST00000177283.8
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr5_-_121511476 | 0.29 |
ENSMUST00000202064.2
|
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr13_-_56283331 | 0.29 |
ENSMUST00000045788.9
ENSMUST00000016081.13 |
Macroh2a1
|
macroH2A.1 histone |
| chr16_-_95387444 | 0.28 |
ENSMUST00000233269.2
|
Erg
|
ETS transcription factor |
| chr10_+_80692948 | 0.28 |
ENSMUST00000220091.2
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr7_+_102090892 | 0.27 |
ENSMUST00000033283.10
|
Rrm1
|
ribonucleotide reductase M1 |
| chr11_-_96714950 | 0.27 |
ENSMUST00000169828.8
ENSMUST00000126949.8 |
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr15_+_41694317 | 0.27 |
ENSMUST00000166917.3
ENSMUST00000230127.2 ENSMUST00000230131.2 |
Oxr1
|
oxidation resistance 1 |
| chr15_+_44291470 | 0.26 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr11_-_45943138 | 0.26 |
ENSMUST00000093169.3
|
Gm12166
|
predicted gene 12166 |
| chr7_-_16651107 | 0.26 |
ENSMUST00000173139.2
|
Calm3
|
calmodulin 3 |
| chr9_-_39405284 | 0.26 |
ENSMUST00000077757.7
ENSMUST00000215065.3 ENSMUST00000216316.3 |
Olfr44
|
olfactory receptor 44 |
| chr7_-_118304930 | 0.25 |
ENSMUST00000207323.2
ENSMUST00000038791.15 |
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr11_+_69792642 | 0.25 |
ENSMUST00000177138.8
ENSMUST00000108617.10 ENSMUST00000177476.8 ENSMUST00000061837.11 |
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr13_-_94383040 | 0.25 |
ENSMUST00000153558.8
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr7_+_140659672 | 0.25 |
ENSMUST00000066873.5
ENSMUST00000163041.2 |
Pkp3
|
plakophilin 3 |
| chr11_-_59727752 | 0.25 |
ENSMUST00000156837.2
|
Cops3
|
COP9 signalosome subunit 3 |
| chr13_-_99027544 | 0.24 |
ENSMUST00000109399.9
|
Tnpo1
|
transportin 1 |
| chr12_-_73093953 | 0.24 |
ENSMUST00000050029.8
|
Six1
|
sine oculis-related homeobox 1 |
| chr3_+_108164242 | 0.24 |
ENSMUST00000090569.10
|
Psma5
|
proteasome subunit alpha 5 |
| chr12_+_76353835 | 0.23 |
ENSMUST00000220321.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr3_+_109481223 | 0.23 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chr16_+_48692976 | 0.22 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr19_+_8875459 | 0.22 |
ENSMUST00000096246.5
ENSMUST00000235274.2 |
Ganab
|
alpha glucosidase 2 alpha neutral subunit |
| chrX_-_153999440 | 0.22 |
ENSMUST00000026318.15
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr1_+_177272297 | 0.22 |
ENSMUST00000193440.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr1_+_163822438 | 0.22 |
ENSMUST00000045694.14
ENSMUST00000111490.2 |
Mettl18
|
methyltransferase like 18 |
| chr1_-_165762469 | 0.22 |
ENSMUST00000069609.12
ENSMUST00000111427.9 ENSMUST00000111426.11 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr9_+_51958453 | 0.21 |
ENSMUST00000163153.9
|
Rdx
|
radixin |
| chr10_+_128626772 | 0.21 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr1_+_127234441 | 0.21 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr15_+_79784365 | 0.21 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr7_-_126620211 | 0.21 |
ENSMUST00000202624.2
|
Gm42742
|
predicted gene 42742 |
| chr17_+_35117905 | 0.21 |
ENSMUST00000097342.10
ENSMUST00000013931.12 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr15_-_44291699 | 0.21 |
ENSMUST00000038719.8
|
Nudcd1
|
NudC domain containing 1 |
| chr4_+_97665843 | 0.20 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chr4_-_150087587 | 0.20 |
ENSMUST00000084117.13
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr17_+_8502682 | 0.20 |
ENSMUST00000124023.8
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr7_-_110673269 | 0.20 |
ENSMUST00000163014.2
|
Eif4g2
|
eukaryotic translation initiation factor 4, gamma 2 |
| chr10_+_127257077 | 0.19 |
ENSMUST00000168780.8
|
R3hdm2
|
R3H domain containing 2 |
| chr1_+_177272215 | 0.19 |
ENSMUST00000192851.2
ENSMUST00000193480.2 ENSMUST00000195388.2 |
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr4_-_156281935 | 0.19 |
ENSMUST00000180572.2
|
Agrn
|
agrin |
| chr15_+_82012114 | 0.19 |
ENSMUST00000089174.6
|
Ccdc134
|
coiled-coil domain containing 134 |
| chr6_-_40562700 | 0.19 |
ENSMUST00000177178.2
ENSMUST00000129948.9 ENSMUST00000101491.11 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr1_-_163822336 | 0.18 |
ENSMUST00000097493.10
ENSMUST00000045876.8 |
BC055324
|
cDNA sequence BC055324 |
| chr9_+_13677266 | 0.18 |
ENSMUST00000152532.8
|
Mtmr2
|
myotubularin related protein 2 |
| chrX_+_55500170 | 0.17 |
ENSMUST00000039374.9
ENSMUST00000101553.9 ENSMUST00000186445.7 |
Ints6l
|
integrator complex subunit 6 like |
| chr2_-_84255602 | 0.17 |
ENSMUST00000074262.9
|
Calcrl
|
calcitonin receptor-like |
| chr11_-_49603501 | 0.17 |
ENSMUST00000020624.7
ENSMUST00000145353.8 |
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr3_-_130524024 | 0.17 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr10_-_7539009 | 0.16 |
ENSMUST00000163085.8
ENSMUST00000159917.8 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr8_+_47193275 | 0.16 |
ENSMUST00000207571.3
|
Irf2
|
interferon regulatory factor 2 |
| chr5_-_65694483 | 0.16 |
ENSMUST00000149167.2
|
Smim14
|
small integral membrane protein 14 |
| chr5_-_23821523 | 0.16 |
ENSMUST00000088392.9
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr14_-_52257113 | 0.16 |
ENSMUST00000166169.4
ENSMUST00000226605.2 |
Zfp219
|
zinc finger protein 219 |
| chr17_+_35117438 | 0.16 |
ENSMUST00000114033.9
ENSMUST00000078061.13 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr7_+_127368981 | 0.16 |
ENSMUST00000118865.8
ENSMUST00000061587.13 ENSMUST00000121504.2 |
Orai3
|
ORAI calcium release-activated calcium modulator 3 |
| chr3_-_14676261 | 0.16 |
ENSMUST00000108365.4
|
Rbis
|
ribosomal biogenesis factor |
| chr4_-_109013807 | 0.15 |
ENSMUST00000161363.2
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr1_-_180641099 | 0.15 |
ENSMUST00000159789.2
ENSMUST00000081026.11 |
H3f3a
|
H3.3 histone A |
| chr3_-_146487102 | 0.15 |
ENSMUST00000005164.12
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr11_+_20151406 | 0.15 |
ENSMUST00000020358.12
ENSMUST00000109602.8 ENSMUST00000109601.8 ENSMUST00000163483.2 |
Rab1a
|
RAB1A, member RAS oncogene family |
| chr6_-_67512768 | 0.15 |
ENSMUST00000058178.6
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr2_+_35581905 | 0.15 |
ENSMUST00000065001.12
|
Dab2ip
|
disabled 2 interacting protein |
| chr17_+_8502594 | 0.15 |
ENSMUST00000155364.8
ENSMUST00000046754.15 |
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr16_+_17327076 | 0.14 |
ENSMUST00000232242.2
|
Lztr1
|
leucine-zipper-like transcriptional regulator, 1 |
| chr17_+_64203017 | 0.14 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr8_+_47192911 | 0.14 |
ENSMUST00000208433.2
|
Irf2
|
interferon regulatory factor 2 |
| chrX_-_133483849 | 0.14 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chrX_+_47712676 | 0.14 |
ENSMUST00000177710.2
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr10_-_7539333 | 0.14 |
ENSMUST00000162606.8
|
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
| chr17_-_34250616 | 0.14 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr2_-_127363251 | 0.13 |
ENSMUST00000028850.15
ENSMUST00000103215.11 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr4_-_82768958 | 0.13 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr3_-_143908060 | 0.13 |
ENSMUST00000121112.6
|
Lmo4
|
LIM domain only 4 |
| chr2_-_6889783 | 0.13 |
ENSMUST00000170438.8
ENSMUST00000114924.10 ENSMUST00000114934.11 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chrX_+_36059274 | 0.13 |
ENSMUST00000016463.4
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 |
| chr1_-_182929025 | 0.13 |
ENSMUST00000171366.7
|
Disp1
|
dispatched RND transporter family member 1 |
| chr17_-_36149100 | 0.13 |
ENSMUST00000134978.3
|
Tubb5
|
tubulin, beta 5 class I |
| chr3_+_102965910 | 0.13 |
ENSMUST00000199367.5
ENSMUST00000199049.5 ENSMUST00000197678.5 |
Nras
|
neuroblastoma ras oncogene |
| chr3_-_143908111 | 0.13 |
ENSMUST00000121796.8
|
Lmo4
|
LIM domain only 4 |
| chr17_+_29170924 | 0.13 |
ENSMUST00000153462.8
|
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr11_-_69811347 | 0.13 |
ENSMUST00000108610.8
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_-_59730654 | 0.13 |
ENSMUST00000019517.10
|
Cops3
|
COP9 signalosome subunit 3 |
| chr10_-_90959817 | 0.12 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr2_+_29968596 | 0.12 |
ENSMUST00000045246.8
|
Pkn3
|
protein kinase N3 |
| chr19_+_47056161 | 0.12 |
ENSMUST00000026027.7
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr10_-_128727542 | 0.12 |
ENSMUST00000026408.7
|
Gdf11
|
growth differentiation factor 11 |
| chr6_+_47854138 | 0.12 |
ENSMUST00000061890.8
|
Zfp282
|
zinc finger protein 282 |
| chr14_+_55716377 | 0.12 |
ENSMUST00000226902.2
ENSMUST00000228353.2 |
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr10_+_61516078 | 0.12 |
ENSMUST00000220372.2
ENSMUST00000020285.10 ENSMUST00000219506.2 ENSMUST00000218474.2 |
Sar1a
|
secretion associated Ras related GTPase 1A |
| chr7_-_24997291 | 0.12 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr13_-_115226666 | 0.12 |
ENSMUST00000109226.5
|
Pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chrX_+_73468140 | 0.12 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr14_+_55716302 | 0.12 |
ENSMUST00000226168.2
|
Dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr16_-_95792090 | 0.12 |
ENSMUST00000233128.2
ENSMUST00000023630.16 |
Psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr19_-_42768374 | 0.12 |
ENSMUST00000069298.13
ENSMUST00000160455.8 ENSMUST00000162004.8 |
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr11_+_102326167 | 0.12 |
ENSMUST00000177428.2
|
Grn
|
granulin |
| chrX_+_47712614 | 0.12 |
ENSMUST00000114936.8
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr7_-_16761732 | 0.11 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr3_-_51184895 | 0.11 |
ENSMUST00000108051.8
ENSMUST00000108053.9 |
Elf2
|
E74-like factor 2 |
| chr1_-_45542442 | 0.11 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr13_-_99027482 | 0.11 |
ENSMUST00000179301.8
ENSMUST00000179271.2 |
Tnpo1
|
transportin 1 |
| chr4_+_88640042 | 0.11 |
ENSMUST00000181601.2
|
Gm26566
|
predicted gene, 26566 |
| chr13_-_47168286 | 0.11 |
ENSMUST00000052747.4
|
Nhlrc1
|
NHL repeat containing 1 |
| chr3_-_108629594 | 0.11 |
ENSMUST00000029482.16
|
Gpsm2
|
G-protein signalling modulator 2 (AGS3-like, C. elegans) |
| chr17_+_29171386 | 0.11 |
ENSMUST00000118762.9
ENSMUST00000057174.16 ENSMUST00000232874.2 ENSMUST00000232772.2 ENSMUST00000233334.2 ENSMUST00000150858.2 ENSMUST00000233064.2 |
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr6_+_51500881 | 0.11 |
ENSMUST00000049152.15
|
Snx10
|
sorting nexin 10 |
| chr16_+_33614378 | 0.11 |
ENSMUST00000115044.8
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr11_+_50116145 | 0.10 |
ENSMUST00000041725.14
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr8_-_27618643 | 0.10 |
ENSMUST00000033877.6
|
Brf2
|
BRF2, RNA polymerase III transcription initiation factor 50kDa subunit |
| chr5_-_36771074 | 0.10 |
ENSMUST00000132383.6
ENSMUST00000174019.2 |
D5Ertd579e
Gm42936
|
DNA segment, Chr 5, ERATO Doi 579, expressed predicted gene 42936 |
| chr1_+_179788675 | 0.10 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr7_+_27305271 | 0.10 |
ENSMUST00000051356.12
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr9_+_103182352 | 0.10 |
ENSMUST00000035164.10
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr11_-_74787877 | 0.10 |
ENSMUST00000057631.12
ENSMUST00000081799.6 |
Sgsm2
|
small G protein signaling modulator 2 |
| chr7_+_27305222 | 0.10 |
ENSMUST00000143499.8
ENSMUST00000108343.8 |
Akt2
|
thymoma viral proto-oncogene 2 |
| chr7_-_19595221 | 0.10 |
ENSMUST00000014830.8
|
Ceacam16
|
carcinoembryonic antigen-related cell adhesion molecule 16 |
| chr7_+_25380195 | 0.10 |
ENSMUST00000205658.2
|
B9d2
|
B9 protein domain 2 |
| chr11_+_3239868 | 0.10 |
ENSMUST00000094471.10
ENSMUST00000110043.8 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr7_-_46365108 | 0.10 |
ENSMUST00000006956.9
ENSMUST00000210913.2 |
Saa3
|
serum amyloid A 3 |
| chr17_+_29538157 | 0.09 |
ENSMUST00000114699.9
ENSMUST00000155348.3 ENSMUST00000234618.2 |
Pi16
|
peptidase inhibitor 16 |
| chr3_-_148696155 | 0.09 |
ENSMUST00000196526.5
ENSMUST00000200543.5 ENSMUST00000200154.5 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr6_-_51443602 | 0.09 |
ENSMUST00000203253.2
|
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr10_-_128383508 | 0.09 |
ENSMUST00000152539.8
ENSMUST00000133458.2 ENSMUST00000040572.10 |
Zc3h10
|
zinc finger CCCH type containing 10 |
| chr17_-_71160477 | 0.09 |
ENSMUST00000118283.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr18_-_39620115 | 0.09 |
ENSMUST00000097592.9
ENSMUST00000115571.8 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr18_-_32692967 | 0.09 |
ENSMUST00000174000.2
ENSMUST00000174459.2 |
Gypc
|
glycophorin C |
| chr3_+_88624194 | 0.09 |
ENSMUST00000172252.2
|
Rit1
|
Ras-like without CAAX 1 |
| chr8_+_72889607 | 0.09 |
ENSMUST00000238492.2
|
Tpm4
|
tropomyosin 4 |
| chr8_-_41469332 | 0.09 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr12_+_86994117 | 0.09 |
ENSMUST00000187814.7
ENSMUST00000038369.11 |
Cipc
|
CLOCK interacting protein, circadian |
| chrX_-_153999333 | 0.09 |
ENSMUST00000112551.4
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr11_+_98689479 | 0.08 |
ENSMUST00000037930.13
|
Msl1
|
male specific lethal 1 |
| chr14_+_70314727 | 0.08 |
ENSMUST00000225200.2
|
Egr3
|
early growth response 3 |
| chr18_+_37847792 | 0.08 |
ENSMUST00000192511.2
ENSMUST00000193476.2 |
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr18_-_36330367 | 0.08 |
ENSMUST00000115713.9
|
Nrg2
|
neuregulin 2 |
| chrX_-_133501677 | 0.08 |
ENSMUST00000239113.2
|
Gla
|
galactosidase, alpha |
| chr6_-_39397334 | 0.08 |
ENSMUST00000031985.13
|
Mkrn1
|
makorin, ring finger protein, 1 |
| chr17_+_71511642 | 0.08 |
ENSMUST00000126681.8
|
Lpin2
|
lipin 2 |
| chr4_+_138161958 | 0.08 |
ENSMUST00000044058.11
ENSMUST00000105813.8 ENSMUST00000105815.2 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr3_+_88624145 | 0.08 |
ENSMUST00000029692.15
ENSMUST00000171645.8 |
Rit1
|
Ras-like without CAAX 1 |
| chrX_-_133501002 | 0.08 |
ENSMUST00000153024.2
|
Gla
|
galactosidase, alpha |
| chr7_-_24997393 | 0.08 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr13_-_74085880 | 0.08 |
ENSMUST00000022053.11
|
Trip13
|
thyroid hormone receptor interactor 13 |
| chr15_+_79784543 | 0.07 |
ENSMUST00000230741.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr9_+_18203421 | 0.07 |
ENSMUST00000001825.9
|
Chordc1
|
cysteine and histidine-rich domain (CHORD)-containing, zinc-binding protein 1 |
| chrX_-_56326951 | 0.07 |
ENSMUST00000114735.9
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr17_-_45903188 | 0.07 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr7_-_24771717 | 0.07 |
ENSMUST00000003468.10
|
Grik5
|
glutamate receptor, ionotropic, kainate 5 (gamma 2) |
| chr12_-_101784727 | 0.07 |
ENSMUST00000222587.2
|
Fbln5
|
fibulin 5 |
| chr7_+_89053562 | 0.07 |
ENSMUST00000058755.5
|
Fzd4
|
frizzled class receptor 4 |
| chr5_-_35259847 | 0.07 |
ENSMUST00000153664.2
|
Lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr6_+_57557978 | 0.07 |
ENSMUST00000031817.10
|
Herc6
|
hect domain and RLD 6 |
| chr3_-_90340830 | 0.07 |
ENSMUST00000029542.12
|
Ints3
|
integrator complex subunit 3 |
| chr11_-_82719850 | 0.07 |
ENSMUST00000021036.13
ENSMUST00000074515.11 ENSMUST00000103218.3 |
Rffl
|
ring finger and FYVE like domain containing protein |
| chr2_+_156038562 | 0.07 |
ENSMUST00000037401.10
|
Phf20
|
PHD finger protein 20 |
| chr5_-_100968795 | 0.07 |
ENSMUST00000117364.8
ENSMUST00000055245.13 |
Abraxas1
|
BRCA1 A complex subunit |
| chrX_-_42256694 | 0.07 |
ENSMUST00000115058.8
ENSMUST00000115059.8 |
Tenm1
|
teneurin transmembrane protein 1 |
| chr10_-_59277570 | 0.07 |
ENSMUST00000009798.5
|
Oit3
|
oncoprotein induced transcript 3 |
| chr18_-_25886750 | 0.07 |
ENSMUST00000224553.2
ENSMUST00000025117.14 |
Celf4
|
CUGBP, Elav-like family member 4 |
| chr4_-_72119070 | 0.07 |
ENSMUST00000102848.9
ENSMUST00000072695.13 ENSMUST00000107337.8 ENSMUST00000074216.14 |
Tle1
|
transducin-like enhancer of split 1 |
| chr9_+_21279802 | 0.07 |
ENSMUST00000214474.2
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr11_+_58197947 | 0.07 |
ENSMUST00000049353.9
|
Zfp692
|
zinc finger protein 692 |
| chr14_-_54843366 | 0.07 |
ENSMUST00000022786.6
|
4931414P19Rik
|
RIKEN cDNA 4931414P19 gene |
| chr14_-_121976273 | 0.06 |
ENSMUST00000212376.3
ENSMUST00000239077.2 |
Dock9
|
dedicator of cytokinesis 9 |
| chr1_-_131441962 | 0.06 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr15_-_79430742 | 0.06 |
ENSMUST00000231053.2
ENSMUST00000229431.2 |
Ddx17
|
DEAD box helicase 17 |
| chr1_-_43866910 | 0.06 |
ENSMUST00000153317.6
ENSMUST00000128261.2 ENSMUST00000126008.8 ENSMUST00000139451.8 |
Uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr1_-_37535202 | 0.06 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr3_-_51248032 | 0.06 |
ENSMUST00000062009.14
ENSMUST00000194641.6 |
Elf2
|
E74-like factor 2 |
| chr2_+_29968225 | 0.06 |
ENSMUST00000150770.8
|
Pkn3
|
protein kinase N3 |
| chr19_-_12742811 | 0.06 |
ENSMUST00000112933.2
|
Cntf
|
ciliary neurotrophic factor |
| chr14_+_60970355 | 0.06 |
ENSMUST00000162945.2
|
Spata13
|
spermatogenesis associated 13 |
| chr3_-_90340910 | 0.06 |
ENSMUST00000196530.2
|
Ints3
|
integrator complex subunit 3 |
| chr8_+_12999394 | 0.06 |
ENSMUST00000110867.9
|
Mcf2l
|
mcf.2 transforming sequence-like |
| chr15_-_88618391 | 0.06 |
ENSMUST00000109381.9
|
Brd1
|
bromodomain containing 1 |
| chr13_-_103901010 | 0.05 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr2_-_28511941 | 0.05 |
ENSMUST00000028156.8
ENSMUST00000164290.8 |
Gfi1b
|
growth factor independent 1B |
| chr1_-_75293447 | 0.05 |
ENSMUST00000189551.7
|
Dnpep
|
aspartyl aminopeptidase |
| chr2_-_52225146 | 0.05 |
ENSMUST00000075301.10
|
Neb
|
nebulin |
| chr15_-_79430942 | 0.05 |
ENSMUST00000054014.9
ENSMUST00000229877.2 |
Ddx17
|
DEAD box helicase 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.3 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.4 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 0.7 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.4 | GO:0036166 | phenotypic switching(GO:0036166) cellular response to cocaine(GO:0071314) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.3 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.2 | GO:0000105 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.2 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 1.0 | GO:0007379 | segment specification(GO:0007379) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.1 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.4 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:1990428 | miRNA transport(GO:1990428) |
| 0.0 | 0.2 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:2000299 | negative regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000299) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.4 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) negative regulation of defense response to virus by host(GO:0050689) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.3 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.2 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.2 | GO:0018455 | alcohol dehydrogenase [NAD(P)+] activity(GO:0018455) |
| 0.1 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.2 | GO:0052692 | raffinose alpha-galactosidase activity(GO:0052692) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |