Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
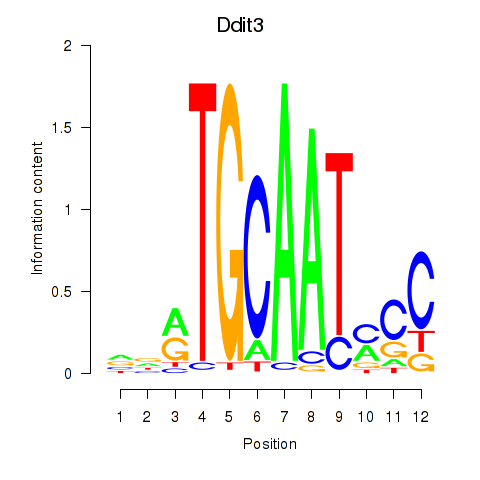
Results for Ddit3
Z-value: 1.98
Transcription factors associated with Ddit3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ddit3
|
ENSMUSG00000025408.16 | DNA-damage inducible transcript 3 |
|
Ddit3
|
ENSMUSG00000116429.2 | DNA-damage inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ddit3 | mm39_v1_chr10_+_127126643_127126698 | -0.51 | 3.8e-01 | Click! |
Activity profile of Ddit3 motif
Sorted Z-values of Ddit3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_67106037 | 2.95 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr7_-_125681577 | 2.51 |
ENSMUST00000073935.7
|
Gsg1l
|
GSG1-like |
| chr19_+_8975249 | 1.91 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr1_+_173501215 | 1.91 |
ENSMUST00000085876.12
|
Ifi208
|
interferon activated gene 208 |
| chr18_+_39126178 | 1.47 |
ENSMUST00000097593.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr1_-_5089564 | 1.45 |
ENSMUST00000002533.15
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr7_-_42376441 | 1.31 |
ENSMUST00000166837.2
|
Gm17067
|
predicted gene 17067 |
| chr13_+_47347301 | 1.27 |
ENSMUST00000110111.4
|
Rnf144b
|
ring finger protein 144B |
| chr3_-_152045986 | 1.23 |
ENSMUST00000199397.2
ENSMUST00000199334.5 ENSMUST00000068243.11 ENSMUST00000073089.13 |
Miga1
|
mitoguardin 1 |
| chr2_-_111843053 | 1.15 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr8_+_127025265 | 1.09 |
ENSMUST00000108759.3
|
Slc35f3
|
solute carrier family 35, member F3 |
| chr18_-_60724855 | 1.05 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr11_-_102446947 | 1.02 |
ENSMUST00000143842.2
|
Gpatch8
|
G patch domain containing 8 |
| chr2_+_36456674 | 0.96 |
ENSMUST00000215879.2
|
Olfr344
|
olfactory receptor 344 |
| chr18_+_39126325 | 0.94 |
ENSMUST00000137497.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr18_-_35760260 | 0.94 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr19_-_4384029 | 0.92 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr1_+_59296065 | 0.87 |
ENSMUST00000160662.8
ENSMUST00000114248.3 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr2_+_36263531 | 0.86 |
ENSMUST00000072114.4
ENSMUST00000217511.2 |
Olfr338
|
olfactory receptor 338 |
| chr16_+_24540599 | 0.86 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr14_-_104081119 | 0.84 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr15_-_76093256 | 0.82 |
ENSMUST00000071869.12
ENSMUST00000170915.2 |
Plec
|
plectin |
| chr2_+_90229027 | 0.79 |
ENSMUST00000216111.3
|
Olfr1274
|
olfactory receptor 1274 |
| chr15_-_58953838 | 0.77 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr7_+_84178162 | 0.75 |
ENSMUST00000180387.3
|
Gm2115
|
predicted gene 2115 |
| chr4_-_56990306 | 0.75 |
ENSMUST00000053681.6
|
Frrs1l
|
ferric-chelate reductase 1 like |
| chr19_-_12147438 | 0.74 |
ENSMUST00000207679.3
ENSMUST00000219261.2 |
Olfr1555-ps1
|
olfactory receptor 1555, pseudogene 1 |
| chr9_+_18848418 | 0.73 |
ENSMUST00000218385.2
|
Olfr832
|
olfactory receptor 832 |
| chr2_-_90054837 | 0.73 |
ENSMUST00000213994.3
|
Olfr1506
|
olfactory receptor 1506 |
| chrX_+_7439839 | 0.72 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr16_+_4964849 | 0.70 |
ENSMUST00000165810.2
ENSMUST00000230616.2 |
Sec14l5
|
SEC14-like lipid binding 5 |
| chr11_-_95966477 | 0.69 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) |
| chr2_-_45007407 | 0.68 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chrY_-_7169293 | 0.68 |
ENSMUST00000189201.2
|
Gm21244
|
predicted gene, 21244 |
| chr9_-_108305683 | 0.67 |
ENSMUST00000076592.4
|
Iho1
|
interactor of HORMAD1 1 |
| chr2_+_181632922 | 0.65 |
ENSMUST00000071760.8
ENSMUST00000236373.2 ENSMUST00000184507.3 |
Gm14496
|
predicted gene 14496 |
| chr8_+_85807566 | 0.63 |
ENSMUST00000140621.2
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr12_-_84240781 | 0.63 |
ENSMUST00000110294.2
|
Mideas
|
mitotic deacetylase associated SANT domain protein |
| chr11_+_101473062 | 0.62 |
ENSMUST00000039581.14
ENSMUST00000100403.9 ENSMUST00000107194.8 ENSMUST00000128614.2 |
Tmem106a
|
transmembrane protein 106A |
| chr10_-_95158827 | 0.54 |
ENSMUST00000220279.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr11_+_70104929 | 0.54 |
ENSMUST00000094055.10
ENSMUST00000126296.8 ENSMUST00000136328.2 ENSMUST00000153993.3 |
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr2_-_172296662 | 0.50 |
ENSMUST00000161334.2
|
Gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr8_+_85807369 | 0.48 |
ENSMUST00000079764.14
|
Wdr83os
|
WD repeat domain 83 opposite strand |
| chr9_+_38312994 | 0.46 |
ENSMUST00000214648.3
ENSMUST00000056364.3 |
Olfr147
|
olfactory receptor 147 |
| chr17_+_26633794 | 0.45 |
ENSMUST00000182897.8
ENSMUST00000183077.8 ENSMUST00000053020.8 |
Neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr5_-_31684036 | 0.45 |
ENSMUST00000202421.2
ENSMUST00000201769.4 ENSMUST00000065388.11 |
Supt7l
|
SPT7-like, STAGA complex gamma subunit |
| chr17_-_5468938 | 0.43 |
ENSMUST00000189788.2
|
Ldhal6b
|
lactate dehydrogenase A-like 6B |
| chr11_-_78136767 | 0.43 |
ENSMUST00000002121.5
|
Supt6
|
SPT6, histone chaperone and transcription elongation factor |
| chr3_+_10431961 | 0.43 |
ENSMUST00000029049.7
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr2_+_90152544 | 0.42 |
ENSMUST00000214797.3
|
Olfr1565
|
olfactory receptor 1565 |
| chr6_+_17463819 | 0.41 |
ENSMUST00000140070.8
|
Met
|
met proto-oncogene |
| chr15_+_59246134 | 0.40 |
ENSMUST00000227173.2
ENSMUST00000079703.11 |
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chrM_+_2743 | 0.39 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr5_-_131567526 | 0.38 |
ENSMUST00000161374.8
|
Auts2
|
autism susceptibility candidate 2 |
| chr15_-_43733389 | 0.38 |
ENSMUST00000067469.6
|
Tmem74
|
transmembrane protein 74 |
| chr11_+_95152355 | 0.37 |
ENSMUST00000021242.5
|
Tac4
|
tachykinin 4 |
| chr2_-_130681603 | 0.35 |
ENSMUST00000119422.8
|
4930402H24Rik
|
RIKEN cDNA 4930402H24 gene |
| chr2_+_30176418 | 0.35 |
ENSMUST00000138666.8
ENSMUST00000113634.3 |
Nup188
|
nucleoporin 188 |
| chr9_+_38725910 | 0.35 |
ENSMUST00000213164.2
|
Olfr922
|
olfactory receptor 922 |
| chrX_+_105186789 | 0.34 |
ENSMUST00000040065.4
|
Tlr13
|
toll-like receptor 13 |
| chr9_+_20556088 | 0.34 |
ENSMUST00000162303.8
ENSMUST00000161486.8 |
Ubl5
|
ubiquitin-like 5 |
| chr6_+_68247469 | 0.33 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr9_+_72345801 | 0.33 |
ENSMUST00000184604.8
ENSMUST00000034746.10 |
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr7_-_44541787 | 0.33 |
ENSMUST00000208551.2
ENSMUST00000208253.2 ENSMUST00000207654.2 ENSMUST00000207278.2 |
Med25
|
mediator complex subunit 25 |
| chr5_-_53864595 | 0.32 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr4_-_118774662 | 0.31 |
ENSMUST00000071979.2
|
Olfr1329
|
olfactory receptor 1329 |
| chr7_-_104677667 | 0.31 |
ENSMUST00000215899.2
ENSMUST00000214318.3 |
Olfr675
|
olfactory receptor 675 |
| chr9_-_45817666 | 0.31 |
ENSMUST00000161187.8
|
Rnf214
|
ring finger protein 214 |
| chr7_+_18618605 | 0.31 |
ENSMUST00000032573.8
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr5_-_5315968 | 0.31 |
ENSMUST00000115451.8
ENSMUST00000115452.8 ENSMUST00000131392.8 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr11_-_118180895 | 0.30 |
ENSMUST00000144153.8
|
Usp36
|
ubiquitin specific peptidase 36 |
| chr11_+_70104736 | 0.29 |
ENSMUST00000171032.8
|
Slc16a11
|
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
| chr19_+_10872778 | 0.28 |
ENSMUST00000179297.3
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr18_+_42408418 | 0.28 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr9_+_13298301 | 0.27 |
ENSMUST00000159294.9
|
Maml2
|
mastermind like transcriptional coactivator 2 |
| chr19_-_12209960 | 0.27 |
ENSMUST00000207710.3
|
Olfr1432
|
olfactory receptor 1432 |
| chr5_-_53864874 | 0.27 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr7_-_23206631 | 0.27 |
ENSMUST00000227713.2
|
Vmn1r167
|
vomeronasal 1 receptor 167 |
| chr15_+_32244947 | 0.27 |
ENSMUST00000067458.7
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr10_+_84753480 | 0.26 |
ENSMUST00000038523.15
ENSMUST00000214693.2 ENSMUST00000095385.5 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr7_-_104628324 | 0.25 |
ENSMUST00000217091.2
ENSMUST00000210963.3 |
Olfr671
|
olfactory receptor 671 |
| chr11_+_98700613 | 0.25 |
ENSMUST00000169695.2
|
Casc3
|
cancer susceptibility candidate 3 |
| chr5_-_92511136 | 0.24 |
ENSMUST00000077820.6
|
Cxcl11
|
chemokine (C-X-C motif) ligand 11 |
| chr3_-_127202693 | 0.24 |
ENSMUST00000182078.9
|
Ank2
|
ankyrin 2, brain |
| chr6_+_45036970 | 0.24 |
ENSMUST00000114641.8
|
Cntnap2
|
contactin associated protein-like 2 |
| chr2_-_120681078 | 0.23 |
ENSMUST00000028740.11
|
Ttbk2
|
tau tubulin kinase 2 |
| chr3_+_60503051 | 0.23 |
ENSMUST00000192757.6
ENSMUST00000193518.6 ENSMUST00000195817.3 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr13_+_24798591 | 0.22 |
ENSMUST00000091694.10
|
Ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr7_+_75292971 | 0.22 |
ENSMUST00000207998.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr3_-_127202663 | 0.22 |
ENSMUST00000182008.8
ENSMUST00000182547.8 |
Ank2
|
ankyrin 2, brain |
| chr19_+_45352514 | 0.21 |
ENSMUST00000224318.2
ENSMUST00000223684.2 |
Btrc
|
beta-transducin repeat containing protein |
| chr2_+_121787131 | 0.20 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr1_-_156766381 | 0.20 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr15_+_59246080 | 0.19 |
ENSMUST00000168722.3
|
Nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr7_+_63937413 | 0.19 |
ENSMUST00000032736.11
|
Mtmr10
|
myotubularin related protein 10 |
| chr8_+_107329983 | 0.19 |
ENSMUST00000000312.12
ENSMUST00000167688.2 |
Cdh1
|
cadherin 1 |
| chr9_-_20556031 | 0.19 |
ENSMUST00000148631.8
ENSMUST00000131128.2 ENSMUST00000151861.9 ENSMUST00000131343.8 ENSMUST00000086458.10 |
Fbxl12
|
F-box and leucine-rich repeat protein 12 |
| chr11_+_110888313 | 0.19 |
ENSMUST00000106635.2
|
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr15_-_83395065 | 0.19 |
ENSMUST00000109479.8
ENSMUST00000109480.8 ENSMUST00000016897.12 |
Ttll1
|
tubulin tyrosine ligase-like 1 |
| chr9_-_40015750 | 0.18 |
ENSMUST00000213858.2
|
Olfr984
|
olfactory receptor 984 |
| chr6_-_114946947 | 0.18 |
ENSMUST00000139640.2
|
Vgll4
|
vestigial like family member 4 |
| chr2_-_35995283 | 0.18 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr4_+_118750282 | 0.18 |
ENSMUST00000094830.2
|
Olfr1330
|
olfactory receptor 1330 |
| chr17_-_35351026 | 0.18 |
ENSMUST00000025249.7
|
Apom
|
apolipoprotein M |
| chr14_+_20398230 | 0.18 |
ENSMUST00000224930.2
ENSMUST00000224110.2 ENSMUST00000225942.2 ENSMUST00000051915.7 ENSMUST00000090499.13 ENSMUST00000224721.2 ENSMUST00000090503.12 ENSMUST00000225991.2 ENSMUST00000037698.13 |
Fam149b
|
family with sequence similarity 149, member B |
| chr18_-_37777238 | 0.17 |
ENSMUST00000066272.6
|
Taf7
|
TATA-box binding protein associated factor 7 |
| chr2_-_88837317 | 0.16 |
ENSMUST00000216271.3
ENSMUST00000214809.3 |
Olfr1215
|
olfactory receptor 1215 |
| chr2_-_148285450 | 0.14 |
ENSMUST00000099269.4
|
Cd93
|
CD93 antigen |
| chr7_-_16549394 | 0.14 |
ENSMUST00000206259.2
|
Fkrp
|
fukutin related protein |
| chr10_-_95159410 | 0.13 |
ENSMUST00000217809.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr7_+_23811739 | 0.13 |
ENSMUST00000120006.8
ENSMUST00000005413.4 |
Zfp112
|
zinc finger protein 112 |
| chr4_+_147953287 | 0.12 |
ENSMUST00000094481.2
|
Fv1
|
Friend virus susceptibility 1 |
| chr9_+_20556147 | 0.12 |
ENSMUST00000161882.8
|
Ubl5
|
ubiquitin-like 5 |
| chr7_+_126376099 | 0.11 |
ENSMUST00000038614.12
ENSMUST00000170882.8 ENSMUST00000106359.2 ENSMUST00000106357.8 ENSMUST00000145762.8 |
Ypel3
|
yippee like 3 |
| chr16_-_45544960 | 0.11 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr3_-_127202586 | 0.11 |
ENSMUST00000183095.3
ENSMUST00000182610.8 |
Ank2
|
ankyrin 2, brain |
| chr11_-_6015736 | 0.11 |
ENSMUST00000002817.12
ENSMUST00000109813.9 ENSMUST00000090443.10 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II, beta |
| chr1_-_36982747 | 0.10 |
ENSMUST00000185964.3
|
Tmem131
|
transmembrane protein 131 |
| chr7_+_120682472 | 0.10 |
ENSMUST00000171880.3
ENSMUST00000047025.15 ENSMUST00000170106.2 ENSMUST00000168311.8 |
Otoa
|
otoancorin |
| chr9_+_65278979 | 0.09 |
ENSMUST00000239433.2
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr2_+_181479647 | 0.09 |
ENSMUST00000029116.14
ENSMUST00000108754.8 |
Pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr13_+_42834039 | 0.09 |
ENSMUST00000128646.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr2_-_32851586 | 0.08 |
ENSMUST00000133832.8
ENSMUST00000124492.8 ENSMUST00000145578.8 ENSMUST00000127321.8 ENSMUST00000113200.8 |
Lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr1_-_156766351 | 0.08 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr7_+_107679062 | 0.08 |
ENSMUST00000213601.2
|
Olfr481
|
olfactory receptor 481 |
| chr4_+_114914607 | 0.06 |
ENSMUST00000136946.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr17_-_15163362 | 0.06 |
ENSMUST00000238668.2
ENSMUST00000228330.2 |
Wdr27
|
WD repeat domain 27 |
| chr9_+_64940004 | 0.06 |
ENSMUST00000167773.2
|
Dpp8
|
dipeptidylpeptidase 8 |
| chr3_+_108093645 | 0.06 |
ENSMUST00000050909.7
ENSMUST00000106659.3 ENSMUST00000106656.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr14_+_26789345 | 0.06 |
ENSMUST00000226105.2
|
Il17rd
|
interleukin 17 receptor D |
| chr6_+_17463748 | 0.06 |
ENSMUST00000115443.8
|
Met
|
met proto-oncogene |
| chr14_+_32972324 | 0.05 |
ENSMUST00000131086.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr5_+_31684331 | 0.05 |
ENSMUST00000114533.9
ENSMUST00000202214.4 ENSMUST00000201858.4 ENSMUST00000202950.4 |
Slc4a1ap
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr18_+_65184052 | 0.05 |
ENSMUST00000224347.3
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr15_+_6673167 | 0.05 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr14_-_70890521 | 0.04 |
ENSMUST00000062629.5
|
Npm2
|
nucleophosmin/nucleoplasmin 2 |
| chr7_+_144450260 | 0.04 |
ENSMUST00000033389.7
ENSMUST00000207229.2 |
Fgf15
|
fibroblast growth factor 15 |
| chr19_+_4889394 | 0.04 |
ENSMUST00000088653.3
|
Ccdc87
|
coiled-coil domain containing 87 |
| chrX_+_165021919 | 0.04 |
ENSMUST00000060210.14
ENSMUST00000112233.8 |
Gpm6b
|
glycoprotein m6b |
| chr4_-_117622747 | 0.03 |
ENSMUST00000062747.6
|
Klf17
|
Kruppel-like factor 17 |
| chr2_+_16361582 | 0.03 |
ENSMUST00000114703.10
|
Plxdc2
|
plexin domain containing 2 |
| chr11_-_103109247 | 0.03 |
ENSMUST00000103076.2
|
Spata32
|
spermatogenesis associated 32 |
| chr11_+_71641806 | 0.03 |
ENSMUST00000108511.8
|
Wscd1
|
WSC domain containing 1 |
| chr3_+_108646974 | 0.02 |
ENSMUST00000133931.9
|
Aknad1
|
AKNA domain containing 1 |
| chr1_+_36753607 | 0.02 |
ENSMUST00000208994.2
|
4933424G06Rik
|
RIKEN cDNA 4933424G06 gene |
| chr17_+_69382694 | 0.02 |
ENSMUST00000225062.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr7_-_79443536 | 0.02 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chr5_-_108922819 | 0.02 |
ENSMUST00000200159.2
ENSMUST00000212212.2 |
Rnf212
|
ring finger protein 212 |
| chr8_-_100143029 | 0.02 |
ENSMUST00000155527.8
ENSMUST00000142129.8 ENSMUST00000093249.11 ENSMUST00000142475.3 ENSMUST00000128860.8 |
Cdh8
|
cadherin 8 |
| chr9_+_57143839 | 0.01 |
ENSMUST00000093837.5
|
Trcg1
|
taste receptor cell gene 1 |
| chr8_+_95113066 | 0.01 |
ENSMUST00000161576.8
ENSMUST00000034220.8 |
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr6_+_90443293 | 0.01 |
ENSMUST00000203607.2
|
Klf15
|
Kruppel-like factor 15 |
| chr5_+_137551774 | 0.01 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr13_-_96028412 | 0.01 |
ENSMUST00000068603.8
|
Iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr6_-_125143425 | 0.01 |
ENSMUST00000117757.9
ENSMUST00000073605.15 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr4_-_118687635 | 0.01 |
ENSMUST00000076019.4
|
Olfr1333
|
olfactory receptor 1333 |
| chr2_-_126718037 | 0.01 |
ENSMUST00000028843.12
|
Trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr16_-_87229367 | 0.01 |
ENSMUST00000232095.2
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr4_-_144190326 | 0.01 |
ENSMUST00000105749.2
|
Aadacl3
|
arylacetamide deacetylase like 3 |
| chr18_+_37466877 | 0.01 |
ENSMUST00000194655.2
ENSMUST00000061717.4 |
Pcdhb6
|
protocadherin beta 6 |
| chr9_-_95727267 | 0.01 |
ENSMUST00000093800.9
|
Pls1
|
plastin 1 (I-isoform) |
| chr3_-_153611775 | 0.01 |
ENSMUST00000005630.11
|
Msh4
|
mutS homolog 4 |
| chr6_-_115569504 | 0.01 |
ENSMUST00000112957.2
|
Mkrn2os
|
makorin, ring finger protein 2, opposite strand |
| chr11_-_114952984 | 0.01 |
ENSMUST00000062787.8
|
Cd300e
|
CD300E molecule |
| chr12_-_87149124 | 0.00 |
ENSMUST00000110176.2
|
Ngb
|
neuroglobin |
| chr15_-_59245998 | 0.00 |
ENSMUST00000022976.6
|
Washc5
|
WASH complex subunit 5 |
| chr1_-_161807205 | 0.00 |
ENSMUST00000162676.2
|
4930558K02Rik
|
RIKEN cDNA 4930558K02 gene |
| chr11_-_59054521 | 0.00 |
ENSMUST00000137433.2
ENSMUST00000054523.6 |
Iba57
|
IBA57 homolog, iron-sulfur cluster assembly |
| chr18_+_37063237 | 0.00 |
ENSMUST00000193839.6
ENSMUST00000070797.7 |
Pcdha1
|
protocadherin alpha 1 |
| chr17_-_15784582 | 0.00 |
ENSMUST00000147532.8
|
Prdm9
|
PR domain containing 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ddit3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 | 0.7 | GO:0002849 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.2 | 1.1 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.8 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.3 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 2.9 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.1 | 0.3 | GO:0051714 | regulation of cytolysis in other organism(GO:0051710) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.9 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 0.6 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.6 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.1 | 0.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.4 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.2 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) |
| 0.1 | 0.5 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.2 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 | 0.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 2.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.4 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0060466 | activation of meiosis involved in egg activation(GO:0060466) |
| 0.0 | 1.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 1.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.7 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0003137 | Notch signaling pathway involved in heart induction(GO:0003137) regulation of Notch signaling pathway involved in heart induction(GO:0035480) positive regulation of Notch signaling pathway involved in heart induction(GO:0035481) |
| 0.0 | 0.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 2.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 2.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 3.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.3 | 0.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 0.9 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 1.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.9 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |