Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Dmc1
Z-value: 0.50
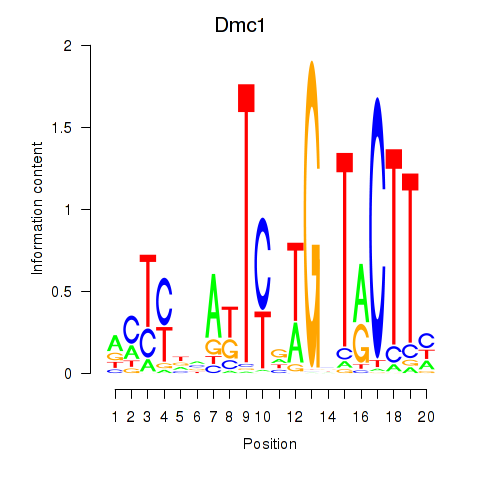
Transcription factors associated with Dmc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dmc1
|
ENSMUSG00000022429.12 | DNA meiotic recombinase 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmc1 | mm39_v1_chr15_-_79489286_79489315 | -0.10 | 8.7e-01 | Click! |
Activity profile of Dmc1 motif
Sorted Z-values of Dmc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_107696793 | 0.29 |
ENSMUST00000217304.2
|
Olfr482
|
olfactory receptor 482 |
| chr5_-_115257336 | 0.26 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr17_+_35643818 | 0.25 |
ENSMUST00000174699.8
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr6_-_7692855 | 0.25 |
ENSMUST00000115542.8
ENSMUST00000148349.2 |
Asns
|
asparagine synthetase |
| chr11_-_29497819 | 0.24 |
ENSMUST00000102844.4
|
Rps27a
|
ribosomal protein S27A |
| chr17_+_35643853 | 0.24 |
ENSMUST00000113879.4
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr4_+_102617332 | 0.23 |
ENSMUST00000066824.14
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr6_-_68713748 | 0.23 |
ENSMUST00000183936.2
ENSMUST00000196863.2 |
Igkv19-93
|
immunoglobulin kappa chain variable 19-93 |
| chr10_+_128745214 | 0.22 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr11_+_3280401 | 0.21 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chrX_+_133657312 | 0.21 |
ENSMUST00000081834.10
ENSMUST00000086880.11 ENSMUST00000086884.5 |
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr2_-_91274967 | 0.19 |
ENSMUST00000064652.14
ENSMUST00000102594.11 ENSMUST00000094835.9 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr3_+_108164242 | 0.19 |
ENSMUST00000090569.10
|
Psma5
|
proteasome subunit alpha 5 |
| chr2_+_103800459 | 0.19 |
ENSMUST00000111143.8
ENSMUST00000138815.2 |
Lmo2
|
LIM domain only 2 |
| chr14_-_73613385 | 0.19 |
ENSMUST00000227454.2
|
Itm2b
|
integral membrane protein 2B |
| chr4_-_42856771 | 0.18 |
ENSMUST00000107981.2
|
Gm12394
|
predicted gene 12394 |
| chr6_+_17749169 | 0.18 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr4_-_107928567 | 0.18 |
ENSMUST00000106701.2
|
Scp2
|
sterol carrier protein 2, liver |
| chr15_+_84208915 | 0.18 |
ENSMUST00000023074.9
|
Parvg
|
parvin, gamma |
| chr6_-_126916487 | 0.17 |
ENSMUST00000144954.5
ENSMUST00000112220.8 ENSMUST00000112221.8 |
Rad51ap1
|
RAD51 associated protein 1 |
| chr13_+_33187205 | 0.17 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr5_+_149202157 | 0.17 |
ENSMUST00000200806.4
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr2_+_84810802 | 0.16 |
ENSMUST00000028467.6
|
Prg2
|
proteoglycan 2, bone marrow |
| chr4_+_138926577 | 0.16 |
ENSMUST00000145368.8
|
Capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr6_-_7693184 | 0.16 |
ENSMUST00000031766.12
|
Asns
|
asparagine synthetase |
| chr7_-_125090540 | 0.16 |
ENSMUST00000138616.3
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr12_-_85420684 | 0.16 |
ENSMUST00000222585.2
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chrX_-_72502595 | 0.16 |
ENSMUST00000033737.15
ENSMUST00000077243.5 |
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr16_-_56984137 | 0.15 |
ENSMUST00000231733.2
|
Nit2
|
nitrilase family, member 2 |
| chr2_+_151385797 | 0.15 |
ENSMUST00000142271.2
|
Fkbp1a
|
FK506 binding protein 1a |
| chr14_-_56026266 | 0.15 |
ENSMUST00000168716.8
ENSMUST00000178399.3 ENSMUST00000022830.14 |
Ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr1_-_75101826 | 0.15 |
ENSMUST00000152855.3
|
Nhej1
|
non-homologous end joining factor 1 |
| chrX_+_134786600 | 0.14 |
ENSMUST00000180025.8
ENSMUST00000148374.8 ENSMUST00000068755.14 |
Bhlhb9
|
basic helix-loop-helix domain containing, class B9 |
| chr17_+_34062059 | 0.14 |
ENSMUST00000002379.15
|
Cd320
|
CD320 antigen |
| chr7_-_79765042 | 0.14 |
ENSMUST00000206714.2
ENSMUST00000107384.10 |
Idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr8_-_57003828 | 0.14 |
ENSMUST00000134162.8
ENSMUST00000140107.8 ENSMUST00000040330.15 ENSMUST00000135337.8 |
Cep44
|
centrosomal protein 44 |
| chr5_-_92496730 | 0.14 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr11_-_48836975 | 0.14 |
ENSMUST00000104958.2
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr17_-_35827676 | 0.14 |
ENSMUST00000160885.2
ENSMUST00000159009.2 ENSMUST00000161012.8 |
Tcf19
|
transcription factor 19 |
| chr11_+_3280771 | 0.13 |
ENSMUST00000136536.8
ENSMUST00000093399.11 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_+_100829798 | 0.13 |
ENSMUST00000106980.9
|
Trim45
|
tripartite motif-containing 45 |
| chr6_+_48653047 | 0.13 |
ENSMUST00000054050.5
|
Gimap9
|
GTPase, IMAP family member 9 |
| chr15_-_60696790 | 0.13 |
ENSMUST00000100635.5
|
Lratd2
|
LRAT domain containing 1 |
| chr8_-_85620537 | 0.13 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr6_-_115652831 | 0.13 |
ENSMUST00000112949.8
|
Raf1
|
v-raf-leukemia viral oncogene 1 |
| chr1_-_192946359 | 0.13 |
ENSMUST00000161737.8
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr12_+_100076407 | 0.13 |
ENSMUST00000021595.10
|
Psmc1
|
protease (prosome, macropain) 26S subunit, ATPase 1 |
| chr4_-_116664729 | 0.13 |
ENSMUST00000106455.8
ENSMUST00000030451.10 |
Toe1
|
target of EGR1, member 1 (nuclear) |
| chr9_+_110867807 | 0.13 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr1_-_52271455 | 0.12 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr3_+_100829750 | 0.12 |
ENSMUST00000037409.13
|
Trim45
|
tripartite motif-containing 45 |
| chr19_-_4675352 | 0.12 |
ENSMUST00000224707.2
ENSMUST00000237059.2 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr5_+_149201577 | 0.12 |
ENSMUST00000071130.5
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr13_+_62277700 | 0.12 |
ENSMUST00000221772.2
ENSMUST00000099449.4 |
Zfp808
|
zinc finger protein 808 |
| chr1_-_184578057 | 0.12 |
ENSMUST00000068725.10
|
Mtarc2
|
mitochondrial amidoxime reducing component 2 |
| chr3_+_14706781 | 0.12 |
ENSMUST00000029071.9
|
Car13
|
carbonic anhydrase 13 |
| chr12_-_79239022 | 0.12 |
ENSMUST00000161204.8
|
Rdh11
|
retinol dehydrogenase 11 |
| chr11_-_6217718 | 0.12 |
ENSMUST00000004507.11
ENSMUST00000151446.2 |
Ddx56
|
DEAD box helicase 56 |
| chr1_+_52026296 | 0.12 |
ENSMUST00000168302.8
|
Stat4
|
signal transducer and activator of transcription 4 |
| chr2_+_103799873 | 0.12 |
ENSMUST00000123437.8
|
Lmo2
|
LIM domain only 2 |
| chr11_-_116164928 | 0.12 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr1_-_156501860 | 0.12 |
ENSMUST00000188964.7
ENSMUST00000190607.2 ENSMUST00000079625.11 |
Tor3a
|
torsin family 3, member A |
| chr19_+_8931187 | 0.12 |
ENSMUST00000096239.7
|
Tut1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr4_-_116508842 | 0.12 |
ENSMUST00000030455.15
|
Akr1a1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr10_-_81201642 | 0.12 |
ENSMUST00000020456.5
|
4930404N11Rik
|
RIKEN cDNA 4930404N11 gene |
| chr6_+_82923876 | 0.11 |
ENSMUST00000113980.4
|
M1ap
|
meiosis 1 associated protein |
| chr18_+_32970363 | 0.11 |
ENSMUST00000166214.9
|
Wdr36
|
WD repeat domain 36 |
| chr17_-_25104874 | 0.11 |
ENSMUST00000234597.2
|
Nubp2
|
nucleotide binding protein 2 |
| chr2_-_126342551 | 0.11 |
ENSMUST00000129187.2
|
Atp8b4
|
ATPase, class I, type 8B, member 4 |
| chr10_-_75673175 | 0.11 |
ENSMUST00000220440.2
|
Gstt2
|
glutathione S-transferase, theta 2 |
| chr11_+_115455260 | 0.11 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr6_-_128414616 | 0.11 |
ENSMUST00000151796.3
|
Fkbp4
|
FK506 binding protein 4 |
| chr17_-_65901946 | 0.11 |
ENSMUST00000232686.2
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr17_-_80514725 | 0.11 |
ENSMUST00000234696.2
ENSMUST00000235069.2 ENSMUST00000063417.11 |
Srsf7
|
serine and arginine-rich splicing factor 7 |
| chr6_+_126916956 | 0.11 |
ENSMUST00000201617.2
|
D6Wsu163e
|
DNA segment, Chr 6, Wayne State University 163, expressed |
| chr17_-_28736483 | 0.11 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr14_-_78866714 | 0.11 |
ENSMUST00000228362.2
ENSMUST00000227767.2 |
Dgkh
|
diacylglycerol kinase, eta |
| chr18_+_77032080 | 0.11 |
ENSMUST00000026485.15
ENSMUST00000150990.9 ENSMUST00000148955.3 |
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr17_+_37581103 | 0.11 |
ENSMUST00000038580.7
|
H2-M3
|
histocompatibility 2, M region locus 3 |
| chr6_+_34361153 | 0.11 |
ENSMUST00000038383.14
ENSMUST00000115051.8 |
Akr1b10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr18_+_32970278 | 0.11 |
ENSMUST00000053663.11
|
Wdr36
|
WD repeat domain 36 |
| chrX_-_149223883 | 0.11 |
ENSMUST00000163233.2
|
Tmem29
|
transmembrane protein 29 |
| chr7_+_46496929 | 0.10 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr19_-_4675300 | 0.10 |
ENSMUST00000225264.2
ENSMUST00000237022.2 ENSMUST00000224675.3 |
Rce1
|
Ras converting CAAX endopeptidase 1 |
| chr1_-_106687457 | 0.10 |
ENSMUST00000010049.6
|
Kdsr
|
3-ketodihydrosphingosine reductase |
| chr19_+_36325683 | 0.10 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr9_+_113641615 | 0.10 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr12_+_103354915 | 0.10 |
ENSMUST00000101094.9
ENSMUST00000021620.13 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr1_+_16735401 | 0.10 |
ENSMUST00000177501.2
ENSMUST00000065373.6 |
Tmem70
|
transmembrane protein 70 |
| chr4_-_117039809 | 0.10 |
ENSMUST00000065896.9
|
Kif2c
|
kinesin family member 2C |
| chr15_-_38518406 | 0.10 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr9_-_37568570 | 0.10 |
ENSMUST00000117654.3
|
Tbrg1
|
transforming growth factor beta regulated gene 1 |
| chr7_+_90091937 | 0.10 |
ENSMUST00000061767.5
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr8_-_123939480 | 0.10 |
ENSMUST00000000759.9
|
Chmp1a
|
charged multivesicular body protein 1A |
| chr19_-_3956991 | 0.10 |
ENSMUST00000126070.9
ENSMUST00000001801.11 |
Tcirg1
|
T cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 |
| chr15_+_82984402 | 0.10 |
ENSMUST00000078218.11
|
Serhl
|
serine hydrolase-like |
| chr7_+_131162338 | 0.10 |
ENSMUST00000208571.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr9_-_21421458 | 0.10 |
ENSMUST00000213167.2
ENSMUST00000034698.9 |
Tmed1
|
transmembrane p24 trafficking protein 1 |
| chr15_+_82984481 | 0.10 |
ENSMUST00000167389.9
ENSMUST00000166427.8 |
Serhl
|
serine hydrolase-like |
| chr11_-_58059293 | 0.10 |
ENSMUST00000172035.8
ENSMUST00000035604.13 ENSMUST00000102711.9 |
Gemin5
|
gem nuclear organelle associated protein 5 |
| chr16_-_96244794 | 0.09 |
ENSMUST00000113773.2
ENSMUST00000000161.14 |
Itgb2l
|
integrin beta 2-like |
| chr14_-_14389372 | 0.09 |
ENSMUST00000023924.4
|
Rpp14
|
ribonuclease P 14 subunit |
| chr7_-_24423715 | 0.09 |
ENSMUST00000081657.6
|
Lypd11
|
Ly6/PLAUR domain containing 11 |
| chr18_-_46874611 | 0.09 |
ENSMUST00000035648.6
|
Atg12
|
autophagy related 12 |
| chr3_-_97517472 | 0.09 |
ENSMUST00000029730.5
|
Chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr16_+_48692976 | 0.09 |
ENSMUST00000065666.6
|
Retnlg
|
resistin like gamma |
| chr16_-_16950241 | 0.09 |
ENSMUST00000023453.10
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr6_+_137731526 | 0.09 |
ENSMUST00000203216.3
ENSMUST00000087675.9 ENSMUST00000203693.3 |
Dera
|
deoxyribose-phosphate aldolase (putative) |
| chr11_+_85202990 | 0.09 |
ENSMUST00000127717.2
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr7_+_126575781 | 0.09 |
ENSMUST00000206450.2
ENSMUST00000205830.2 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr19_-_38031774 | 0.09 |
ENSMUST00000226068.2
|
Myof
|
myoferlin |
| chr5_+_115417725 | 0.09 |
ENSMUST00000040421.11
|
Coq5
|
coenzyme Q5 methyltransferase |
| chr2_-_25436884 | 0.09 |
ENSMUST00000114234.2
|
Traf2
|
TNF receptor-associated factor 2 |
| chr6_-_125208738 | 0.09 |
ENSMUST00000043422.8
|
Tapbpl
|
TAP binding protein-like |
| chr7_+_65343156 | 0.09 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr17_+_25517363 | 0.09 |
ENSMUST00000037453.4
|
Prss34
|
protease, serine 34 |
| chr7_-_117728790 | 0.09 |
ENSMUST00000206491.2
|
Arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr2_-_66086919 | 0.09 |
ENSMUST00000125446.3
ENSMUST00000102718.10 |
Ttc21b
|
tetratricopeptide repeat domain 21B |
| chr6_-_129449739 | 0.09 |
ENSMUST00000112076.9
ENSMUST00000184581.3 |
Clec7a
|
C-type lectin domain family 7, member a |
| chr11_+_51858476 | 0.09 |
ENSMUST00000102763.5
|
Cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr9_+_73009680 | 0.09 |
ENSMUST00000034737.13
ENSMUST00000173734.9 ENSMUST00000167514.2 ENSMUST00000174203.3 |
Khdc3
Gm20509
|
KH domain containing 3, subcortical maternal complex member predicted gene 20509 |
| chr19_-_24938909 | 0.09 |
ENSMUST00000025815.10
|
Cbwd1
|
COBW domain containing 1 |
| chr16_-_87237438 | 0.09 |
ENSMUST00000039101.12
|
Rwdd2b
|
RWD domain containing 2B |
| chr13_-_62771948 | 0.08 |
ENSMUST00000223338.2
ENSMUST00000201563.4 |
6720489N17Rik
|
RIKEN cDNA 6720489N17 gene |
| chr15_+_79784365 | 0.08 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr1_+_85992341 | 0.08 |
ENSMUST00000027432.9
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr2_-_26917921 | 0.08 |
ENSMUST00000102890.11
ENSMUST00000153388.2 ENSMUST00000045702.6 |
Slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr7_-_143294051 | 0.08 |
ENSMUST00000119499.8
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr19_-_45548942 | 0.08 |
ENSMUST00000026239.7
|
Poll
|
polymerase (DNA directed), lambda |
| chrX_+_69429475 | 0.08 |
ENSMUST00000053981.6
|
Eola1
|
endothelium and lymphocyte associated ASCH domain 1 |
| chr2_+_35512023 | 0.08 |
ENSMUST00000091010.12
|
Dab2ip
|
disabled 2 interacting protein |
| chr2_-_86109346 | 0.08 |
ENSMUST00000217294.2
ENSMUST00000217245.2 ENSMUST00000216432.2 |
Olfr1051
|
olfactory receptor 1051 |
| chr10_+_81193325 | 0.08 |
ENSMUST00000044844.9
|
Mfsd12
|
major facilitator superfamily domain containing 12 |
| chr1_-_194813843 | 0.08 |
ENSMUST00000075451.12
ENSMUST00000191775.2 |
Cr1l
|
complement component (3b/4b) receptor 1-like |
| chr3_+_96537484 | 0.08 |
ENSMUST00000200647.2
|
Rbm8a
|
RNA binding motif protein 8a |
| chr7_-_46558754 | 0.08 |
ENSMUST00000209538.2
|
Tsg101
|
tumor susceptibility gene 101 |
| chr13_-_62530965 | 0.08 |
ENSMUST00000202194.2
ENSMUST00000187656.7 |
Gm3604
|
predicted gene 3604 |
| chr2_+_83554741 | 0.08 |
ENSMUST00000028499.11
|
Itgav
|
integrin alpha V |
| chr8_+_124204598 | 0.08 |
ENSMUST00000001520.13
|
Afg3l1
|
AFG3-like AAA ATPase 1 |
| chr11_-_4799345 | 0.08 |
ENSMUST00000053079.13
ENSMUST00000109910.9 |
Nf2
|
neurofibromin 2 |
| chr7_-_30325514 | 0.08 |
ENSMUST00000208838.2
|
Cox6b1
|
cytochrome c oxidase, subunit 6B1 |
| chr13_+_30320446 | 0.08 |
ENSMUST00000047311.16
|
Mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr17_-_66756710 | 0.08 |
ENSMUST00000086693.12
ENSMUST00000097291.10 |
Mtcl1
|
microtubule crosslinking factor 1 |
| chr12_+_59060162 | 0.08 |
ENSMUST00000021379.8
|
Gemin2
|
gem nuclear organelle associated protein 2 |
| chr9_+_104930438 | 0.08 |
ENSMUST00000149243.8
ENSMUST00000035177.15 ENSMUST00000214036.2 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr9_+_21504018 | 0.08 |
ENSMUST00000062125.11
|
Timm29
|
translocase of inner mitochondrial membrane 29 |
| chr4_+_130001349 | 0.08 |
ENSMUST00000030563.6
|
Pef1
|
penta-EF hand domain containing 1 |
| chr2_-_119617985 | 0.08 |
ENSMUST00000110793.8
ENSMUST00000099529.9 ENSMUST00000048493.12 |
Rpap1
|
RNA polymerase II associated protein 1 |
| chr2_-_152857239 | 0.08 |
ENSMUST00000028972.9
|
Pdrg1
|
p53 and DNA damage regulated 1 |
| chr1_-_75118436 | 0.08 |
ENSMUST00000189403.7
|
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr17_-_31731222 | 0.08 |
ENSMUST00000236665.2
|
Wdr4
|
WD repeat domain 4 |
| chr12_+_55445560 | 0.07 |
ENSMUST00000021412.9
|
Psma6
|
proteasome subunit alpha 6 |
| chr16_-_10360893 | 0.07 |
ENSMUST00000184863.8
ENSMUST00000038281.6 |
Dexi
|
dexamethasone-induced transcript |
| chr12_+_84332006 | 0.07 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chrX_+_105059305 | 0.07 |
ENSMUST00000033582.5
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr6_+_29272625 | 0.07 |
ENSMUST00000054445.9
|
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr8_-_85807308 | 0.07 |
ENSMUST00000093357.12
|
Wdr83
|
WD repeat domain containing 83 |
| chr10_+_61516078 | 0.07 |
ENSMUST00000220372.2
ENSMUST00000020285.10 ENSMUST00000219506.2 ENSMUST00000218474.2 |
Sar1a
|
secretion associated Ras related GTPase 1A |
| chr10_-_17898977 | 0.07 |
ENSMUST00000020002.9
|
Abracl
|
ABRA C-terminal like |
| chr7_-_4066125 | 0.07 |
ENSMUST00000108600.9
ENSMUST00000205296.2 |
Lair1
|
leukocyte-associated Ig-like receptor 1 |
| chr7_-_27374017 | 0.07 |
ENSMUST00000036453.14
ENSMUST00000108341.2 |
Map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr7_-_23998735 | 0.07 |
ENSMUST00000145131.8
|
Zfp61
|
zinc finger protein 61 |
| chr14_+_21881794 | 0.07 |
ENSMUST00000152562.8
|
Vdac2
|
voltage-dependent anion channel 2 |
| chr7_-_4066194 | 0.07 |
ENSMUST00000086400.13
|
Lair1
|
leukocyte-associated Ig-like receptor 1 |
| chr5_-_104169785 | 0.07 |
ENSMUST00000031251.16
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr15_-_38518458 | 0.07 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr2_+_176115387 | 0.07 |
ENSMUST00000108995.9
ENSMUST00000127687.8 |
Zfp968-ps
|
zinc finger protein 968, pseudogene |
| chr7_+_126575752 | 0.07 |
ENSMUST00000206346.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr16_-_65359406 | 0.07 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr5_+_45650821 | 0.07 |
ENSMUST00000198534.2
|
Lap3
|
leucine aminopeptidase 3 |
| chr4_-_11254253 | 0.07 |
ENSMUST00000044616.10
ENSMUST00000108319.9 ENSMUST00000108318.3 |
Ints8
|
integrator complex subunit 8 |
| chr4_+_11558905 | 0.07 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr15_-_103123711 | 0.07 |
ENSMUST00000122182.2
ENSMUST00000108813.10 ENSMUST00000127191.2 |
Cbx5
|
chromobox 5 |
| chr1_+_170846482 | 0.07 |
ENSMUST00000078825.5
|
Fcgr4
|
Fc receptor, IgG, low affinity IV |
| chr15_-_75781138 | 0.07 |
ENSMUST00000145764.2
ENSMUST00000116440.9 ENSMUST00000151066.8 |
Eef1d
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr8_-_26505605 | 0.07 |
ENSMUST00000016138.11
|
Fnta
|
farnesyltransferase, CAAX box, alpha |
| chr9_-_56151334 | 0.07 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr4_-_49521036 | 0.07 |
ENSMUST00000057829.4
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
| chr2_+_173579285 | 0.07 |
ENSMUST00000067530.6
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C |
| chr1_-_58735106 | 0.07 |
ENSMUST00000055313.14
ENSMUST00000188772.7 |
Flacc1
|
flagellum associated containing coiled-coil domains 1 |
| chr9_-_99450948 | 0.07 |
ENSMUST00000035043.12
|
Armc8
|
armadillo repeat containing 8 |
| chr7_-_90106375 | 0.07 |
ENSMUST00000032844.7
|
Tmem126a
|
transmembrane protein 126A |
| chr18_-_35795233 | 0.06 |
ENSMUST00000025209.12
ENSMUST00000096573.4 |
Spata24
|
spermatogenesis associated 24 |
| chr1_-_75118335 | 0.06 |
ENSMUST00000190679.7
|
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr3_+_135531548 | 0.06 |
ENSMUST00000167390.8
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8 |
| chr4_-_128512401 | 0.06 |
ENSMUST00000074829.4
|
Tlr12
|
toll-like receptor 12 |
| chr5_-_107745443 | 0.06 |
ENSMUST00000100949.10
ENSMUST00000078021.13 |
Glmn
|
glomulin, FKBP associated protein |
| chr4_-_117539431 | 0.06 |
ENSMUST00000102687.4
|
Dmap1
|
DNA methyltransferase 1-associated protein 1 |
| chr17_+_26895379 | 0.06 |
ENSMUST00000236299.2
ENSMUST00000167352.2 |
Atp6v0e
|
ATPase, H+ transporting, lysosomal V0 subunit E |
| chr8_+_46984016 | 0.06 |
ENSMUST00000152423.2
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1 |
| chr12_+_86994117 | 0.06 |
ENSMUST00000187814.7
ENSMUST00000038369.11 |
Cipc
|
CLOCK interacting protein, circadian |
| chr4_-_133615075 | 0.06 |
ENSMUST00000003741.16
ENSMUST00000105894.11 |
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_+_6511133 | 0.06 |
ENSMUST00000160633.8
ENSMUST00000109721.3 |
Ccm2
|
cerebral cavernous malformation 2 |
| chr1_-_72323464 | 0.06 |
ENSMUST00000027381.13
|
Pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr5_-_115622356 | 0.06 |
ENSMUST00000112067.8
|
Sirt4
|
sirtuin 4 |
| chr11_-_72106418 | 0.06 |
ENSMUST00000021157.9
|
Med31
|
mediator complex subunit 31 |
| chr11_+_114566257 | 0.06 |
ENSMUST00000045779.6
|
Ttyh2
|
tweety family member 2 |
| chr5_+_122347792 | 0.06 |
ENSMUST00000072602.14
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr1_-_120197979 | 0.06 |
ENSMUST00000112639.8
|
Steap3
|
STEAP family member 3 |
| chr6_-_82629835 | 0.06 |
ENSMUST00000160281.8
ENSMUST00000095786.6 |
Pole4
|
polymerase (DNA-directed), epsilon 4 (p12 subunit) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dmc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.2 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.2 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.2 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.3 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:1904464 | regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) |
| 0.0 | 0.3 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.2 | GO:0032385 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.0 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.0 | GO:0090265 | immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.2 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.1 | GO:1903719 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.0 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.0 | GO:0002654 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.0 | GO:0032077 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0070358 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.1 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.0 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.0 | 0.0 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.0 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0019659 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.3 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.0 | 0.1 | GO:1903754 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0031597 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:1904121 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.0 | 0.1 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0030942 | signal recognition particle binding(GO:0005047) endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.0 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.0 | GO:0045142 | triplex DNA binding(GO:0045142) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.0 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |