|
chr13_+_21938258
Show fit
|
3.40 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15
|
|
chr13_+_22220000
Show fit
|
3.24 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12
|
|
chr13_+_21971631
Show fit
|
3.09 |
ENSMUST00000110473.3
ENSMUST00000102982.2
|
H2bc22
|
H2B clustered histone 22
|
|
chr13_+_22227359
Show fit
|
2.73 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11
|
|
chr13_+_22017906
Show fit
|
2.55 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24
|
|
chr13_-_21900313
Show fit
|
2.30 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13
|
|
chr13_-_23929490
Show fit
|
2.30 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3
|
|
chr13_-_23758370
Show fit
|
1.90 |
ENSMUST00000105106.2
|
H2bc7
|
H2B clustered histone 7
|
|
chr13_-_23746543
Show fit
|
1.73 |
ENSMUST00000105107.2
|
H3c6
|
H3 clustered histone 6
|
|
chr13_-_22219738
Show fit
|
1.72 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12
|
|
chr13_-_21967540
Show fit
|
1.61 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11
|
|
chr13_-_21937997
Show fit
|
1.60 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15
|
|
chr13_-_21994366
Show fit
|
1.59 |
ENSMUST00000091749.4
|
H2bc23
|
H2B clustered histone 23
|
|
chr13_-_21971388
Show fit
|
1.59 |
ENSMUST00000091751.3
|
H2ac22
|
H2A clustered histone 22
|
|
chr3_+_96154001
Show fit
|
1.58 |
ENSMUST00000176059.2
ENSMUST00000177796.2
|
H3c14
|
H3 clustered histone 14
|
|
chr13_-_22017677
Show fit
|
1.35 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24
|
|
chr18_+_10617858
Show fit
|
1.32 |
ENSMUST00000235020.2
|
Snrpd1
|
small nuclear ribonucleoprotein D1
|
|
chr13_-_22227114
Show fit
|
1.30 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11
|
|
chr13_+_21906214
Show fit
|
0.95 |
ENSMUST00000224651.2
|
H2bc14
|
H2B clustered histone 14
|
|
chr13_+_21994588
Show fit
|
0.95 |
ENSMUST00000091745.6
|
H2ac23
|
H2A clustered histone 23
|
|
chr7_-_48530777
Show fit
|
0.94 |
ENSMUST00000058745.15
|
E2f8
|
E2F transcription factor 8
|
|
chr10_+_80633865
Show fit
|
0.93 |
ENSMUST00000148665.8
|
Sf3a2
|
splicing factor 3a, subunit 2
|
|
chr7_-_48531344
Show fit
|
0.88 |
ENSMUST00000119223.2
|
E2f8
|
E2F transcription factor 8
|
|
chr13_+_21900554
Show fit
|
0.88 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13
|
|
chr7_-_45045097
Show fit
|
0.85 |
ENSMUST00000211121.2
ENSMUST00000074575.11
|
Snrnp70
|
small nuclear ribonucleoprotein 70 (U1)
|
|
chr6_-_38852857
Show fit
|
0.85 |
ENSMUST00000162359.8
|
Hipk2
|
homeodomain interacting protein kinase 2
|
|
chr9_+_100525807
Show fit
|
0.84 |
ENSMUST00000133388.2
|
Stag1
|
stromal antigen 1
|
|
chr12_+_11316101
Show fit
|
0.84 |
ENSMUST00000218866.2
|
Smc6
|
structural maintenance of chromosomes 6
|
|
chr3_+_97920819
Show fit
|
0.81 |
ENSMUST00000079812.8
|
Notch2
|
notch 2
|
|
chr11_+_50492899
Show fit
|
0.77 |
ENSMUST00000142118.3
ENSMUST00000040523.9
|
Adamts2
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2
|
|
chr19_-_6134703
Show fit
|
0.70 |
ENSMUST00000161548.8
|
Zfpl1
|
zinc finger like protein 1
|
|
chr10_+_110581293
Show fit
|
0.66 |
ENSMUST00000174857.8
ENSMUST00000073781.12
ENSMUST00000173471.8
ENSMUST00000173634.2
|
E2f7
|
E2F transcription factor 7
|
|
chr8_-_123404811
Show fit
|
0.66 |
ENSMUST00000006525.14
ENSMUST00000064674.13
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3
|
|
chr10_-_60055082
Show fit
|
0.64 |
ENSMUST00000135158.9
|
Chst3
|
carbohydrate sulfotransferase 3
|
|
chr4_+_135899678
Show fit
|
0.59 |
ENSMUST00000061721.6
|
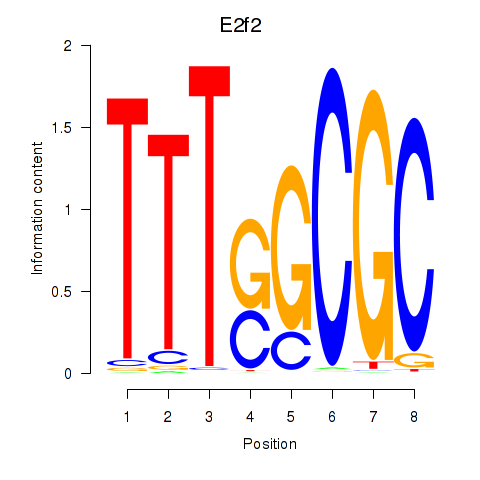
E2f2
|
E2F transcription factor 2
|
|
chr10_+_80634488
Show fit
|
0.58 |
ENSMUST00000151928.8
|
Sf3a2
|
splicing factor 3a, subunit 2
|
|
chr9_-_21003268
Show fit
|
0.57 |
ENSMUST00000115487.3
|
Raver1
|
ribonucleoprotein, PTB-binding 1
|
|
chr9_+_21458138
Show fit
|
0.57 |
ENSMUST00000034703.15
ENSMUST00000115395.10
ENSMUST00000115394.8
|
Carm1
|
coactivator-associated arginine methyltransferase 1
|
|
chr16_-_18066591
Show fit
|
0.56 |
ENSMUST00000115645.10
|
Ranbp1
|
RAN binding protein 1
|
|
chrX_+_41239872
Show fit
|
0.56 |
ENSMUST00000123245.8
|
Stag2
|
stromal antigen 2
|
|
chr5_-_106844396
Show fit
|
0.55 |
ENSMUST00000137285.8
ENSMUST00000124263.2
ENSMUST00000112695.4
ENSMUST00000155495.8
ENSMUST00000135108.2
ENSMUST00000149128.3
|
Zfp644
Gm28039
|
zinc finger protein 644
predicted gene, 28039
|
|
chr9_+_100525501
Show fit
|
0.54 |
ENSMUST00000146312.8
ENSMUST00000129269.8
|
Stag1
|
stromal antigen 1
|
|
chr18_+_10617773
Show fit
|
0.54 |
ENSMUST00000002551.5
ENSMUST00000234207.2
|
Snrpd1
|
small nuclear ribonucleoprotein D1
|
|
chr2_-_180351778
Show fit
|
0.54 |
ENSMUST00000103057.8
ENSMUST00000103055.8
|
Dido1
|
death inducer-obliterator 1
|
|
chr7_+_126380655
Show fit
|
0.53 |
ENSMUST00000172352.8
ENSMUST00000094037.5
|
Tbx6
|
T-box 6
|
|
chr6_-_38853097
Show fit
|
0.52 |
ENSMUST00000161779.8
|
Hipk2
|
homeodomain interacting protein kinase 2
|
|
chr5_-_143846600
Show fit
|
0.52 |
ENSMUST00000031613.11
ENSMUST00000100483.3
|
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2
|
|
chr1_+_176642226
Show fit
|
0.49 |
ENSMUST00000056773.15
ENSMUST00000027785.15
|
Sdccag8
|
serologically defined colon cancer antigen 8
|
|
chr11_-_102446947
Show fit
|
0.49 |
ENSMUST00000143842.2
|
Gpatch8
|
G patch domain containing 8
|
|
chr15_-_98465516
Show fit
|
0.49 |
ENSMUST00000012104.7
|
Ccnt1
|
cyclin T1
|
|
chr4_+_84802592
Show fit
|
0.48 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein
|
|
chr19_-_7194912
Show fit
|
0.47 |
ENSMUST00000039758.6
|
Cox8a
|
cytochrome c oxidase subunit 8A
|
|
chr17_-_33979280
Show fit
|
0.46 |
ENSMUST00000173860.8
|
Rab11b
|
RAB11B, member RAS oncogene family
|
|
chr19_+_53588808
Show fit
|
0.46 |
ENSMUST00000025930.10
|
Smc3
|
structural maintenance of chromosomes 3
|
|
chrX_+_93278203
Show fit
|
0.46 |
ENSMUST00000153900.8
|
Klhl15
|
kelch-like 15
|
|
chrX_+_150799414
Show fit
|
0.45 |
ENSMUST00000045312.6
|
Smc1a
|
structural maintenance of chromosomes 1A
|
|
chrX_+_162691978
Show fit
|
0.45 |
ENSMUST00000069041.15
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr9_+_100525637
Show fit
|
0.45 |
ENSMUST00000041418.13
|
Stag1
|
stromal antigen 1
|
|
chr11_+_58839716
Show fit
|
0.43 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2
|
|
chr19_+_23736205
Show fit
|
0.41 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1
|
|
chr4_+_126450762
Show fit
|
0.40 |
ENSMUST00000147675.2
|
Clspn
|
claspin
|
|
chr13_+_23758555
Show fit
|
0.40 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7
|
|
chr14_-_73563212
Show fit
|
0.39 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1
|
|
chrX_+_93278588
Show fit
|
0.39 |
ENSMUST00000096369.10
ENSMUST00000113911.9
|
Klhl15
|
kelch-like 15
|
|
chr4_-_135080437
Show fit
|
0.39 |
ENSMUST00000030613.11
ENSMUST00000131373.8
|
Srrm1
|
serine/arginine repetitive matrix 1
|
|
chr6_-_149003171
Show fit
|
0.39 |
ENSMUST00000111557.8
|
Dennd5b
|
DENN/MADD domain containing 5B
|
|
chr2_-_30364219
Show fit
|
0.39 |
ENSMUST00000065134.4
|
Ier5l
|
immediate early response 5-like
|
|
chr5_-_106844685
Show fit
|
0.37 |
ENSMUST00000127434.8
ENSMUST00000112696.8
ENSMUST00000112698.8
|
Zfp644
|
zinc finger protein 644
|
|
chr9_+_13739003
Show fit
|
0.37 |
ENSMUST00000059579.12
|
Fam76b
|
family with sequence similarity 76, member B
|
|
chr1_-_176641607
Show fit
|
0.37 |
ENSMUST00000195717.6
ENSMUST00000192961.6
|
Cep170
|
centrosomal protein 170
|
|
chr2_-_32271833
Show fit
|
0.37 |
ENSMUST00000146423.2
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene
|
|
chr14_+_27150750
Show fit
|
0.35 |
ENSMUST00000022450.6
|
Tasor
|
transcription activation suppressor
|
|
chr10_-_125225298
Show fit
|
0.35 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7
|
|
chr17_-_26727437
Show fit
|
0.35 |
ENSMUST00000236661.2
ENSMUST00000025025.7
|
Dusp1
|
dual specificity phosphatase 1
|
|
chr6_-_149003003
Show fit
|
0.34 |
ENSMUST00000127727.2
|
Dennd5b
|
DENN/MADD domain containing 5B
|
|
chrX_+_47430221
Show fit
|
0.34 |
ENSMUST00000136348.8
|
Bcorl1
|
BCL6 co-repressor-like 1
|
|
chr16_+_51851917
Show fit
|
0.34 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr10_-_80634293
Show fit
|
0.34 |
ENSMUST00000218209.2
ENSMUST00000036805.7
|
Plekhj1
|
pleckstrin homology domain containing, family J member 1
|
|
chr16_-_76170714
Show fit
|
0.34 |
ENSMUST00000231585.2
ENSMUST00000121927.8
|
Nrip1
|
nuclear receptor interacting protein 1
|
|
chr1_+_11484313
Show fit
|
0.33 |
ENSMUST00000171690.9
ENSMUST00000048613.14
|
A830018L16Rik
|
RIKEN cDNA A830018L16 gene
|
|
chr16_-_90081300
Show fit
|
0.32 |
ENSMUST00000039280.9
ENSMUST00000232371.2
|
Scaf4
|
SR-related CTD-associated factor 4
|
|
chrX_+_137815171
Show fit
|
0.32 |
ENSMUST00000064937.14
ENSMUST00000113052.8
|
Nrk
|
Nik related kinase
|
|
chr15_-_58953838
Show fit
|
0.32 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1
|
|
chr12_+_33197692
Show fit
|
0.31 |
ENSMUST00000077456.13
ENSMUST00000110824.9
|
Atxn7l1
|
ataxin 7-like 1
|
|
chrX_+_162692126
Show fit
|
0.31 |
ENSMUST00000033734.14
ENSMUST00000112294.9
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr2_+_127267069
Show fit
|
0.31 |
ENSMUST00000062211.4
|
Gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial
|
|
chr16_-_10131804
Show fit
|
0.31 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2
|
|
chr11_-_59730569
Show fit
|
0.31 |
ENSMUST00000141415.2
|
Cops3
|
COP9 signalosome subunit 3
|
|
chr2_-_32271921
Show fit
|
0.30 |
ENSMUST00000048792.5
|
1110008P14Rik
|
RIKEN cDNA 1110008P14 gene
|
|
chr7_-_25454177
Show fit
|
0.30 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr12_+_33197776
Show fit
|
0.30 |
ENSMUST00000125192.9
ENSMUST00000136644.8
|
Atxn7l1
|
ataxin 7-like 1
|
|
chr2_-_45000389
Show fit
|
0.30 |
ENSMUST00000201804.4
ENSMUST00000028229.13
ENSMUST00000202187.4
ENSMUST00000153561.6
ENSMUST00000201490.2
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr2_-_26012751
Show fit
|
0.30 |
ENSMUST00000140993.2
ENSMUST00000028300.6
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing
|
|
chr2_-_45000250
Show fit
|
0.30 |
ENSMUST00000201211.4
ENSMUST00000177302.8
|
Zeb2
|
zinc finger E-box binding homeobox 2
|
|
chr11_-_62349334
Show fit
|
0.29 |
ENSMUST00000141447.2
|
Ncor1
|
nuclear receptor co-repressor 1
|
|
chr11_+_77353218
Show fit
|
0.29 |
ENSMUST00000102493.8
|
Coro6
|
coronin 6
|
|
chr16_-_90081188
Show fit
|
0.29 |
ENSMUST00000163419.9
|
Scaf4
|
SR-related CTD-associated factor 4
|
|
chr4_+_132766793
Show fit
|
0.29 |
ENSMUST00000105914.2
|
Ahdc1
|
AT hook, DNA binding motif, containing 1
|
|
chr4_-_135080471
Show fit
|
0.29 |
ENSMUST00000084846.12
ENSMUST00000136342.9
ENSMUST00000105861.8
|
Srrm1
|
serine/arginine repetitive matrix 1
|
|
chr2_+_91480513
Show fit
|
0.28 |
ENSMUST00000090614.11
|
Arhgap1
|
Rho GTPase activating protein 1
|
|
chr12_+_33197753
Show fit
|
0.28 |
ENSMUST00000146040.9
|
Atxn7l1
|
ataxin 7-like 1
|
|
chr3_-_95125051
Show fit
|
0.26 |
ENSMUST00000107204.8
|
Gabpb2
|
GA repeat binding protein, beta 2
|
|
chr5_+_137786031
Show fit
|
0.26 |
ENSMUST00000035852.14
|
Zcwpw1
|
zinc finger, CW type with PWWP domain 1
|
|
chr12_-_72455708
Show fit
|
0.26 |
ENSMUST00000078505.14
|
Rtn1
|
reticulon 1
|
|
chr11_+_62349238
Show fit
|
0.26 |
ENSMUST00000014389.6
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L
|
|
chr19_+_6413840
Show fit
|
0.26 |
ENSMUST00000113488.8
ENSMUST00000113487.8
|
Sf1
|
splicing factor 1
|
|
chr8_+_83891972
Show fit
|
0.26 |
ENSMUST00000034145.11
|
Tbc1d9
|
TBC1 domain family, member 9
|
|
chr1_-_86287080
Show fit
|
0.25 |
ENSMUST00000027438.8
|
Ncl
|
nucleolin
|
|
chr8_+_84874654
Show fit
|
0.25 |
ENSMUST00000143833.8
ENSMUST00000118856.8
|
Brme1
|
break repair meiotic recombinase recruitment factor 1
|
|
chr6_+_8259288
Show fit
|
0.25 |
ENSMUST00000159335.8
|
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1
|
|
chr9_-_102231884
Show fit
|
0.25 |
ENSMUST00000035129.14
ENSMUST00000085169.12
ENSMUST00000149800.3
|
Ephb1
|
Eph receptor B1
|
|
chr7_+_44465714
Show fit
|
0.24 |
ENSMUST00000208172.2
|
Nup62
|
nucleoporin 62
|
|
chr1_-_64776890
Show fit
|
0.24 |
ENSMUST00000116133.4
ENSMUST00000063982.7
|
Fzd5
|
frizzled class receptor 5
|
|
chr17_+_74835417
Show fit
|
0.24 |
ENSMUST00000182944.8
ENSMUST00000182597.8
ENSMUST00000182133.8
ENSMUST00000183224.8
|
Birc6
|
baculoviral IAP repeat-containing 6
|
|
chr15_-_97991114
Show fit
|
0.24 |
ENSMUST00000180657.2
|
Senp1
|
SUMO1/sentrin specific peptidase 1
|
|
chr17_-_36568728
Show fit
|
0.24 |
ENSMUST00000077535.5
|
Rpp21
|
ribonuclease P 21 subunit
|
|
chr6_+_119152210
Show fit
|
0.24 |
ENSMUST00000112777.9
ENSMUST00000073909.6
|
Dcp1b
|
decapping mRNA 1B
|
|
chr8_+_69333143
Show fit
|
0.24 |
ENSMUST00000015712.15
|
Lpl
|
lipoprotein lipase
|
|
chr11_-_101875575
Show fit
|
0.23 |
ENSMUST00000176261.2
ENSMUST00000143177.2
ENSMUST00000003612.13
|
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related)
|
|
chr2_+_69553384
Show fit
|
0.23 |
ENSMUST00000040915.15
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G)
|
|
chr15_+_30172716
Show fit
|
0.23 |
ENSMUST00000081728.7
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2
|
|
chr1_+_131838220
Show fit
|
0.23 |
ENSMUST00000189946.7
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr19_+_6413703
Show fit
|
0.23 |
ENSMUST00000131252.8
ENSMUST00000113489.8
|
Sf1
|
splicing factor 1
|
|
chr7_-_25454126
Show fit
|
0.23 |
ENSMUST00000108401.3
ENSMUST00000043765.14
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr2_+_69553141
Show fit
|
0.23 |
ENSMUST00000090858.10
|
Ppig
|
peptidyl-prolyl isomerase G (cyclophilin G)
|
|
chr19_-_6134903
Show fit
|
0.22 |
ENSMUST00000160977.8
ENSMUST00000159859.2
ENSMUST00000025707.9
ENSMUST00000160712.8
ENSMUST00000237738.2
|
Zfpl1
|
zinc finger like protein 1
|
|
chr15_+_82183143
Show fit
|
0.22 |
ENSMUST00000023089.5
|
Wbp2nl
|
WBP2 N-terminal like
|
|
chr2_+_91480460
Show fit
|
0.21 |
ENSMUST00000111331.9
|
Arhgap1
|
Rho GTPase activating protein 1
|
|
chrX_+_93278526
Show fit
|
0.21 |
ENSMUST00000113908.8
ENSMUST00000113916.10
|
Klhl15
|
kelch-like 15
|
|
chr5_-_136596299
Show fit
|
0.21 |
ENSMUST00000004097.16
|
Cux1
|
cut-like homeobox 1
|
|
chr7_+_29931309
Show fit
|
0.21 |
ENSMUST00000019882.16
ENSMUST00000149654.8
|
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I
|
|
chr4_+_99082131
Show fit
|
0.21 |
ENSMUST00000030279.15
|
Atg4c
|
autophagy related 4C, cysteine peptidase
|
|
chr5_-_136596094
Show fit
|
0.21 |
ENSMUST00000175975.9
ENSMUST00000176216.9
ENSMUST00000176745.8
|
Cux1
|
cut-like homeobox 1
|
|
chr13_-_96269076
Show fit
|
0.21 |
ENSMUST00000161263.8
|
Sv2c
|
synaptic vesicle glycoprotein 2c
|
|
chr9_+_44245981
Show fit
|
0.20 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone
|
|
chr8_+_84874881
Show fit
|
0.20 |
ENSMUST00000093375.5
|
Brme1
|
break repair meiotic recombinase recruitment factor 1
|
|
chr17_-_23959334
Show fit
|
0.20 |
ENSMUST00000024702.5
|
Paqr4
|
progestin and adipoQ receptor family member IV
|
|
chr14_-_67106037
Show fit
|
0.20 |
ENSMUST00000022629.9
|
Dpysl2
|
dihydropyrimidinase-like 2
|
|
chr1_-_74544946
Show fit
|
0.19 |
ENSMUST00000044260.11
ENSMUST00000186282.7
|
Usp37
|
ubiquitin specific peptidase 37
|
|
chr18_-_80756273
Show fit
|
0.19 |
ENSMUST00000078049.12
ENSMUST00000236711.2
|
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1
|
|
chr1_-_38168801
Show fit
|
0.19 |
ENSMUST00000195383.2
|
Rev1
|
REV1, DNA directed polymerase
|
|
chr8_-_84874468
Show fit
|
0.19 |
ENSMUST00000117424.9
ENSMUST00000040383.9
|
Cc2d1a
|
coiled-coil and C2 domain containing 1A
|
|
chr3_-_116388334
Show fit
|
0.18 |
ENSMUST00000197190.5
ENSMUST00000198454.2
|
Trmt13
|
tRNA methyltransferase 13
|
|
chr7_+_19093665
Show fit
|
0.18 |
ENSMUST00000140836.8
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr18_-_16942289
Show fit
|
0.18 |
ENSMUST00000025166.14
|
Cdh2
|
cadherin 2
|
|
chr1_+_191307748
Show fit
|
0.18 |
ENSMUST00000045450.7
|
Ints7
|
integrator complex subunit 7
|
|
chr15_+_61857226
Show fit
|
0.18 |
ENSMUST00000161976.8
ENSMUST00000022971.8
|
Myc
|
myelocytomatosis oncogene
|
|
chr9_+_122780111
Show fit
|
0.18 |
ENSMUST00000040717.7
ENSMUST00000214652.2
ENSMUST00000217401.2
|
Kif15
|
kinesin family member 15
|
|
chr2_-_58990967
Show fit
|
0.18 |
ENSMUST00000226455.2
ENSMUST00000077687.6
|
Ccdc148
|
coiled-coil domain containing 148
|
|
chr13_+_54769611
Show fit
|
0.17 |
ENSMUST00000026991.16
ENSMUST00000137413.8
ENSMUST00000135232.8
ENSMUST00000124752.2
|
Faf2
|
Fas associated factor family member 2
|
|
chr18_+_36693024
Show fit
|
0.17 |
ENSMUST00000134146.8
|
Ankhd1
|
ankyrin repeat and KH domain containing 1
|
|
chr15_+_78912642
Show fit
|
0.17 |
ENSMUST00000180086.3
|
H1f0
|
H1.0 linker histone
|
|
chr5_-_25703700
Show fit
|
0.17 |
ENSMUST00000173073.8
ENSMUST00000045291.14
ENSMUST00000173174.2
|
Kmt2c
|
lysine (K)-specific methyltransferase 2C
|
|
chr3_+_116388600
Show fit
|
0.17 |
ENSMUST00000198386.5
ENSMUST00000198311.5
ENSMUST00000197335.2
|
Sass6
|
SAS-6 centriolar assembly protein
|
|
chr1_+_11484489
Show fit
|
0.17 |
ENSMUST00000135014.8
|
A830018L16Rik
|
RIKEN cDNA A830018L16 gene
|
|
chr6_+_4601124
Show fit
|
0.17 |
ENSMUST00000141359.2
|
Casd1
|
CAS1 domain containing 1
|
|
chr1_+_61677977
Show fit
|
0.16 |
ENSMUST00000075374.10
|
Pard3b
|
par-3 family cell polarity regulator beta
|
|
chr7_-_16348862
Show fit
|
0.16 |
ENSMUST00000171937.2
ENSMUST00000075845.11
|
Arhgap35
|
Rho GTPase activating protein 35
|
|
chr15_-_8473918
Show fit
|
0.16 |
ENSMUST00000052965.8
|
Nipbl
|
NIPBL cohesin loading factor
|
|
chr9_+_61280501
Show fit
|
0.16 |
ENSMUST00000162583.8
ENSMUST00000161993.8
ENSMUST00000160882.8
ENSMUST00000160724.8
ENSMUST00000162973.8
ENSMUST00000159050.8
|
Tle3
|
transducin-like enhancer of split 3
|
|
chr2_+_53082079
Show fit
|
0.16 |
ENSMUST00000028336.7
|
Arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6
|
|
chr9_+_61280764
Show fit
|
0.16 |
ENSMUST00000160541.8
ENSMUST00000161207.8
ENSMUST00000159630.8
|
Tle3
|
transducin-like enhancer of split 3
|
|
chrX_+_150586309
Show fit
|
0.16 |
ENSMUST00000026292.15
|
Huwe1
|
HECT, UBA and WWE domain containing 1
|
|
chr8_-_89088782
Show fit
|
0.15 |
ENSMUST00000034085.8
|
Brd7
|
bromodomain containing 7
|
|
chr18_-_35760260
Show fit
|
0.15 |
ENSMUST00000025212.8
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr9_-_63929308
Show fit
|
0.15 |
ENSMUST00000179458.2
ENSMUST00000041029.6
|
Smad6
|
SMAD family member 6
|
|
chr5_-_135518098
Show fit
|
0.15 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1
|
|
chr17_-_35265514
Show fit
|
0.15 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5
|
|
chr2_+_164721277
Show fit
|
0.15 |
ENSMUST00000041643.5
|
Pcif1
|
phosphorylated CTD interacting factor 1
|
|
chr4_+_114857370
Show fit
|
0.14 |
ENSMUST00000129957.8
|
Stil
|
Scl/Tal1 interrupting locus
|
|
chr17_+_50816296
Show fit
|
0.14 |
ENSMUST00000043938.8
|
Plcl2
|
phospholipase C-like 2
|
|
chr15_-_72932853
Show fit
|
0.14 |
ENSMUST00000170633.9
ENSMUST00000228960.2
|
Trappc9
|
trafficking protein particle complex 9
|
|
chr18_-_38068456
Show fit
|
0.14 |
ENSMUST00000080033.7
ENSMUST00000115631.8
ENSMUST00000115634.8
|
Diaph1
|
diaphanous related formin 1
|
|
chr3_-_151543088
Show fit
|
0.14 |
ENSMUST00000029670.7
|
Ptgfr
|
prostaglandin F receptor
|
|
chr4_+_84802513
Show fit
|
0.14 |
ENSMUST00000047023.13
|
Cntln
|
centlein, centrosomal protein
|
|
chr4_+_137196080
Show fit
|
0.14 |
ENSMUST00000030547.15
ENSMUST00000171332.2
|
Hspg2
|
perlecan (heparan sulfate proteoglycan 2)
|
|
chr11_+_115921129
Show fit
|
0.14 |
ENSMUST00000021116.12
ENSMUST00000106452.2
|
Unk
|
unkempt family zinc finger
|
|
chr11_-_77498636
Show fit
|
0.14 |
ENSMUST00000058496.8
|
Taok1
|
TAO kinase 1
|
|
chr9_+_61280890
Show fit
|
0.13 |
ENSMUST00000161689.8
|
Tle3
|
transducin-like enhancer of split 3
|
|
chr7_-_6733411
Show fit
|
0.13 |
ENSMUST00000239104.2
ENSMUST00000051209.11
|
Peg3
|
paternally expressed 3
|
|
chr19_+_46064302
Show fit
|
0.13 |
ENSMUST00000165017.2
ENSMUST00000223741.2
ENSMUST00000225780.2
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr4_+_114857348
Show fit
|
0.13 |
ENSMUST00000030490.13
|
Stil
|
Scl/Tal1 interrupting locus
|
|
chr19_+_6291698
Show fit
|
0.13 |
ENSMUST00000045351.13
|
Atg2a
|
autophagy related 2A
|
|
chr7_+_44465353
Show fit
|
0.13 |
ENSMUST00000208626.2
ENSMUST00000057195.17
|
Nup62
|
nucleoporin 62
|
|
chr7_+_112806672
Show fit
|
0.13 |
ENSMUST00000047321.9
ENSMUST00000210074.2
ENSMUST00000210238.2
|
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr18_-_64622092
Show fit
|
0.13 |
ENSMUST00000235766.3
ENSMUST00000025484.9
ENSMUST00000237502.2
ENSMUST00000236586.3
|
Fech
|
ferrochelatase
|
|
chr15_-_57998443
Show fit
|
0.13 |
ENSMUST00000038194.5
|
Atad2
|
ATPase family, AAA domain containing 2
|
|
chr1_+_183766572
Show fit
|
0.12 |
ENSMUST00000048655.8
|
Dusp10
|
dual specificity phosphatase 10
|
|
chr15_+_9140614
Show fit
|
0.12 |
ENSMUST00000227556.3
|
Lmbrd2
|
LMBR1 domain containing 2
|
|
chr13_-_47259652
Show fit
|
0.12 |
ENSMUST00000021807.13
ENSMUST00000135278.8
|
Dek
|
DEK proto-oncogene (DNA binding)
|
|
chr18_-_40352372
Show fit
|
0.12 |
ENSMUST00000025364.6
|
Yipf5
|
Yip1 domain family, member 5
|
|
chr8_-_70664901
Show fit
|
0.12 |
ENSMUST00000063788.8
ENSMUST00000110127.8
|
Slc25a42
|
solute carrier family 25, member 42
|
|
chr15_+_61857390
Show fit
|
0.12 |
ENSMUST00000159327.2
ENSMUST00000167731.8
|
Myc
|
myelocytomatosis oncogene
|
|
chr4_-_11966367
Show fit
|
0.11 |
ENSMUST00000056050.5
ENSMUST00000108299.2
ENSMUST00000108297.3
|
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr13_-_47259266
Show fit
|
0.11 |
ENSMUST00000129352.3
|
Dek
|
DEK proto-oncogene (DNA binding)
|
|
chr18_+_31937129
Show fit
|
0.11 |
ENSMUST00000082319.15
ENSMUST00000234957.2
ENSMUST00000025264.8
ENSMUST00000234344.2
|
Wdr33
|
WD repeat domain 33
|
|
chr5_+_65921414
Show fit
|
0.11 |
ENSMUST00000201615.4
ENSMUST00000087264.4
|
N4bp2
|
NEDD4 binding protein 2
|
|
chr2_+_121787131
Show fit
|
0.11 |
ENSMUST00000110574.8
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr15_-_79489286
Show fit
|
0.11 |
ENSMUST00000229408.2
ENSMUST00000023065.8
|
Dmc1
|
DNA meiotic recombinase 1
|
|
chr18_-_38068430
Show fit
|
0.11 |
ENSMUST00000025337.14
|
Diaph1
|
diaphanous related formin 1
|