Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for E2f3
Z-value: 1.78
Transcription factors associated with E2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f3
|
ENSMUSG00000016477.19 | E2F transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f3 | mm39_v1_chr13_-_30170031_30170048 | 1.00 | 3.6e-05 | Click! |
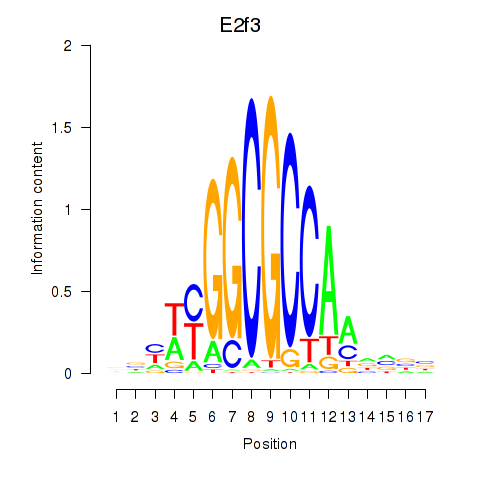
Activity profile of E2f3 motif
Sorted Z-values of E2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_79967543 | 1.47 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr12_+_24758968 | 1.39 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr1_-_91340884 | 1.17 |
ENSMUST00000086851.2
|
Hes6
|
hairy and enhancer of split 6 |
| chr12_+_24758724 | 1.01 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr18_+_31922173 | 0.90 |
ENSMUST00000025106.5
ENSMUST00000234146.2 |
Polr2d
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr6_+_86342622 | 0.87 |
ENSMUST00000071492.9
|
Fam136a
|
family with sequence similarity 136, member A |
| chr5_-_65855511 | 0.85 |
ENSMUST00000201948.4
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr12_+_24758240 | 0.85 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr3_-_84063067 | 0.77 |
ENSMUST00000047368.8
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr10_-_34003933 | 0.75 |
ENSMUST00000062784.8
|
Calhm6
|
calcium homeostasis modulator family member 6 |
| chr4_+_132495636 | 0.72 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr2_+_150751475 | 0.72 |
ENSMUST00000028948.5
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr11_+_115455260 | 0.65 |
ENSMUST00000021085.11
|
Nup85
|
nucleoporin 85 |
| chr2_+_127112613 | 0.62 |
ENSMUST00000125049.2
ENSMUST00000110374.2 |
Stard7
|
START domain containing 7 |
| chr3_+_10077608 | 0.57 |
ENSMUST00000029046.9
|
Fabp5
|
fatty acid binding protein 5, epidermal |
| chr2_+_31560725 | 0.53 |
ENSMUST00000038474.14
ENSMUST00000137156.2 |
Exosc2
|
exosome component 2 |
| chr1_-_66902429 | 0.53 |
ENSMUST00000027153.6
|
Acadl
|
acyl-Coenzyme A dehydrogenase, long-chain |
| chr5_-_100867520 | 0.52 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr18_+_47245204 | 0.49 |
ENSMUST00000234633.2
|
Hspe1-rs1
|
heat shock protein 1 (chaperonin 10), related sequence 1 |
| chr16_-_4536992 | 0.49 |
ENSMUST00000115851.10
|
Nmral1
|
NmrA-like family domain containing 1 |
| chr17_+_29251602 | 0.48 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr16_-_91443794 | 0.48 |
ENSMUST00000232367.2
ENSMUST00000231380.2 ENSMUST00000231444.2 ENSMUST00000232289.2 ENSMUST00000120450.2 ENSMUST00000023684.14 |
Gart
|
phosphoribosylglycinamide formyltransferase |
| chr11_+_21189277 | 0.47 |
ENSMUST00000109578.8
ENSMUST00000006221.14 |
Vps54
|
VPS54 GARP complex subunit |
| chrX_-_101232978 | 0.47 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr3_-_95125190 | 0.47 |
ENSMUST00000136139.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr4_-_108436514 | 0.47 |
ENSMUST00000079213.6
|
Prpf38a
|
PRP38 pre-mRNA processing factor 38 (yeast) domain containing A |
| chr4_+_131570770 | 0.43 |
ENSMUST00000030742.11
ENSMUST00000137321.3 |
Mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr1_-_39616369 | 0.42 |
ENSMUST00000195705.2
|
Rnf149
|
ring finger protein 149 |
| chr6_-_34887743 | 0.42 |
ENSMUST00000081214.12
|
Wdr91
|
WD repeat domain 91 |
| chr3_+_96542933 | 0.40 |
ENSMUST00000048766.15
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr9_+_64188857 | 0.40 |
ENSMUST00000215031.2
ENSMUST00000213165.2 ENSMUST00000213289.2 ENSMUST00000216594.2 ENSMUST00000034964.7 |
Tipin
|
timeless interacting protein |
| chr5_-_123038329 | 0.40 |
ENSMUST00000031435.14
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr8_-_124586159 | 0.40 |
ENSMUST00000034452.12
|
Ccsap
|
centriole, cilia and spindle associated protein |
| chr18_+_74912268 | 0.39 |
ENSMUST00000041053.11
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) |
| chr19_+_3373285 | 0.39 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr17_+_34258411 | 0.39 |
ENSMUST00000087497.11
ENSMUST00000131134.9 ENSMUST00000235819.2 ENSMUST00000114255.9 ENSMUST00000114252.9 ENSMUST00000237989.2 |
Col11a2
|
collagen, type XI, alpha 2 |
| chr1_-_91326490 | 0.39 |
ENSMUST00000190519.2
ENSMUST00000027534.13 ENSMUST00000187306.7 |
Ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase 2C |
| chr8_+_106587268 | 0.38 |
ENSMUST00000212610.2
ENSMUST00000212484.2 ENSMUST00000212200.2 |
Nutf2
|
nuclear transport factor 2 |
| chr10_-_71180763 | 0.38 |
ENSMUST00000045887.9
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr19_+_5416769 | 0.37 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr10_-_60983438 | 0.36 |
ENSMUST00000092498.12
ENSMUST00000137833.2 ENSMUST00000155919.8 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr11_-_101442663 | 0.36 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr1_-_63215812 | 0.35 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr14_-_47805861 | 0.35 |
ENSMUST00000228784.2
ENSMUST00000042988.7 |
Atg14
|
autophagy related 14 |
| chr11_-_84020382 | 0.35 |
ENSMUST00000018795.13
|
Tada2a
|
transcriptional adaptor 2A |
| chr3_+_81839908 | 0.35 |
ENSMUST00000029649.3
|
Ctso
|
cathepsin O |
| chr16_-_4536943 | 0.35 |
ENSMUST00000120056.8
ENSMUST00000074970.8 |
Nmral1
|
NmrA-like family domain containing 1 |
| chr1_-_135241429 | 0.35 |
ENSMUST00000134088.3
ENSMUST00000081104.10 |
Timm17a
|
translocase of inner mitochondrial membrane 17a |
| chr2_-_24864998 | 0.35 |
ENSMUST00000194392.2
|
Mrpl41
|
mitochondrial ribosomal protein L41 |
| chr3_+_96543143 | 0.35 |
ENSMUST00000165842.3
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr3_-_65300000 | 0.35 |
ENSMUST00000029414.12
|
Ssr3
|
signal sequence receptor, gamma |
| chr1_+_156386414 | 0.35 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr7_+_140461860 | 0.34 |
ENSMUST00000026560.14
|
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr3_-_121608859 | 0.34 |
ENSMUST00000029770.8
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr2_+_144398226 | 0.34 |
ENSMUST00000155876.8
ENSMUST00000149697.3 |
Sec23b
|
SEC23 homolog B, COPII coat complex component |
| chr17_+_36152559 | 0.34 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr18_+_11790409 | 0.33 |
ENSMUST00000047322.8
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr10_+_80062468 | 0.33 |
ENSMUST00000130260.2
|
Pwwp3a
|
PWWP domain containing 3A, DNA repair factor |
| chr3_-_65299967 | 0.33 |
ENSMUST00000119896.2
|
Ssr3
|
signal sequence receptor, gamma |
| chr9_-_98483695 | 0.33 |
ENSMUST00000035034.10
|
Mrps22
|
mitochondrial ribosomal protein S22 |
| chr1_-_128287347 | 0.33 |
ENSMUST00000190495.2
ENSMUST00000027601.11 |
Mcm6
|
minichromosome maintenance complex component 6 |
| chr16_-_87229485 | 0.32 |
ENSMUST00000039449.9
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr13_+_54722833 | 0.32 |
ENSMUST00000156024.2
|
Arl10
|
ADP-ribosylation factor-like 10 |
| chr16_-_22258469 | 0.32 |
ENSMUST00000079601.13
|
Etv5
|
ets variant 5 |
| chr5_-_92653377 | 0.32 |
ENSMUST00000031377.9
|
Scarb2
|
scavenger receptor class B, member 2 |
| chr3_-_95125002 | 0.32 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr13_-_21586858 | 0.32 |
ENSMUST00000117721.8
ENSMUST00000070785.16 ENSMUST00000116433.2 ENSMUST00000223831.2 ENSMUST00000116434.11 ENSMUST00000224820.2 |
Zkscan3
|
zinc finger with KRAB and SCAN domains 3 |
| chr7_+_34818709 | 0.32 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr12_+_111505253 | 0.31 |
ENSMUST00000220803.2
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr3_-_94949854 | 0.31 |
ENSMUST00000117355.2
ENSMUST00000071664.12 ENSMUST00000107237.8 |
Psmd4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr15_-_79430942 | 0.31 |
ENSMUST00000054014.9
ENSMUST00000229877.2 |
Ddx17
|
DEAD box helicase 17 |
| chr8_-_57940834 | 0.30 |
ENSMUST00000034022.4
|
Sap30
|
sin3 associated polypeptide |
| chr2_+_144398149 | 0.30 |
ENSMUST00000143573.8
ENSMUST00000028916.15 ENSMUST00000155258.2 |
Sec23b
|
SEC23 homolog B, COPII coat complex component |
| chr3_-_121608809 | 0.29 |
ENSMUST00000197383.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr10_-_80754016 | 0.28 |
ENSMUST00000057623.14
ENSMUST00000179022.8 |
Lmnb2
|
lamin B2 |
| chrX_+_20570145 | 0.27 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr2_-_118987305 | 0.27 |
ENSMUST00000135419.8
ENSMUST00000129351.2 ENSMUST00000139519.2 ENSMUST00000094695.12 |
Rmdn3
|
regulator of microtubule dynamics 3 |
| chr9_+_55949141 | 0.26 |
ENSMUST00000114276.3
|
Rcn2
|
reticulocalbin 2 |
| chr7_-_140462221 | 0.26 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr6_-_72322116 | 0.25 |
ENSMUST00000070345.5
|
Usp39
|
ubiquitin specific peptidase 39 |
| chr2_+_28531239 | 0.25 |
ENSMUST00000028155.12
ENSMUST00000113869.8 ENSMUST00000113867.9 |
Tsc1
|
TSC complex subunit 1 |
| chr8_-_120331936 | 0.25 |
ENSMUST00000093099.13
|
Taf1c
|
TATA-box binding protein associated factor, RNA polymerase I, C |
| chr9_+_63509925 | 0.25 |
ENSMUST00000041551.9
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr8_-_35432783 | 0.25 |
ENSMUST00000033929.6
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr12_-_81468705 | 0.24 |
ENSMUST00000085319.4
|
Adam4
|
a disintegrin and metallopeptidase domain 4 |
| chr3_+_33853941 | 0.24 |
ENSMUST00000099153.10
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr6_+_29768470 | 0.24 |
ENSMUST00000102995.9
ENSMUST00000115242.9 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr16_-_87229367 | 0.23 |
ENSMUST00000232095.2
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr2_+_150628655 | 0.23 |
ENSMUST00000045441.8
|
Pygb
|
brain glycogen phosphorylase |
| chr10_-_30476658 | 0.23 |
ENSMUST00000019927.7
|
Trmt11
|
tRNA methyltransferase 11 |
| chr6_-_71417607 | 0.23 |
ENSMUST00000002292.15
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr13_-_53135064 | 0.22 |
ENSMUST00000071065.8
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr16_-_43709968 | 0.22 |
ENSMUST00000023387.14
|
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr1_-_63215952 | 0.22 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr12_+_24701273 | 0.21 |
ENSMUST00000020982.7
|
Klf11
|
Kruppel-like factor 11 |
| chr2_-_120183575 | 0.21 |
ENSMUST00000028752.8
ENSMUST00000102501.10 |
Vps39
|
VPS39 HOPS complex subunit |
| chr7_-_140462187 | 0.21 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr9_-_79700660 | 0.21 |
ENSMUST00000034878.12
|
Tmem30a
|
transmembrane protein 30A |
| chr9_-_72892617 | 0.20 |
ENSMUST00000124565.3
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr7_+_97049210 | 0.20 |
ENSMUST00000032882.9
ENSMUST00000149122.2 |
Ndufc2
|
NADH:ubiquinone oxidoreductase subunit C2 |
| chr9_-_105372235 | 0.20 |
ENSMUST00000176190.8
ENSMUST00000163879.9 ENSMUST00000112558.10 ENSMUST00000176363.9 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr12_-_69274936 | 0.20 |
ENSMUST00000221411.2
ENSMUST00000021359.7 |
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr3_+_96542746 | 0.20 |
ENSMUST00000118557.8
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr11_+_65698001 | 0.20 |
ENSMUST00000071465.9
ENSMUST00000018491.8 |
Zkscan6
|
zinc finger with KRAB and SCAN domains 6 |
| chr6_-_24528012 | 0.19 |
ENSMUST00000023851.9
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr9_-_20888054 | 0.19 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr7_-_140461769 | 0.19 |
ENSMUST00000106048.10
ENSMUST00000147331.9 ENSMUST00000137710.2 |
Sirt3
|
sirtuin 3 |
| chr13_+_92491234 | 0.19 |
ENSMUST00000022218.6
|
Dhfr
|
dihydrofolate reductase |
| chr10_-_60983404 | 0.18 |
ENSMUST00000122259.8
|
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr17_-_80369626 | 0.18 |
ENSMUST00000184635.8
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr3_+_66893280 | 0.18 |
ENSMUST00000161726.2
ENSMUST00000160504.2 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr17_+_27136065 | 0.18 |
ENSMUST00000078961.6
|
Kifc5b
|
kinesin family member C5B |
| chr17_-_33007238 | 0.18 |
ENSMUST00000159086.10
|
Zfp871
|
zinc finger protein 871 |
| chr1_+_44187007 | 0.18 |
ENSMUST00000027214.4
|
Ercc5
|
excision repair cross-complementing rodent repair deficiency, complementation group 5 |
| chr17_-_71782296 | 0.18 |
ENSMUST00000127430.2
|
Smchd1
|
SMC hinge domain containing 1 |
| chr6_-_88818832 | 0.18 |
ENSMUST00000032169.8
|
Abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr1_-_16689527 | 0.18 |
ENSMUST00000182554.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr1_-_95595280 | 0.17 |
ENSMUST00000043336.11
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr7_-_45362867 | 0.17 |
ENSMUST00000211340.2
|
Sphk2
|
sphingosine kinase 2 |
| chr7_-_100021514 | 0.17 |
ENSMUST00000032963.10
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chrX_+_73326303 | 0.17 |
ENSMUST00000069722.13
ENSMUST00000124200.8 ENSMUST00000114180.11 ENSMUST00000132437.8 |
Taz
|
tafazzin |
| chr4_+_43957677 | 0.16 |
ENSMUST00000107855.2
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr9_+_108731287 | 0.16 |
ENSMUST00000188557.8
|
Slc26a6
|
solute carrier family 26, member 6 |
| chr12_+_85157607 | 0.16 |
ENSMUST00000053811.10
|
Dlst
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| chr7_+_12568647 | 0.16 |
ENSMUST00000004614.15
|
Zfp110
|
zinc finger protein 110 |
| chr1_-_16689660 | 0.16 |
ENSMUST00000117146.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr17_-_80369762 | 0.16 |
ENSMUST00000061331.14
|
Hnrnpll
|
heterogeneous nuclear ribonucleoprotein L-like |
| chr10_-_80512117 | 0.16 |
ENSMUST00000200082.5
|
Mknk2
|
MAP kinase-interacting serine/threonine kinase 2 |
| chr14_-_14255736 | 0.16 |
ENSMUST00000170111.3
|
Kctd6
|
potassium channel tetramerisation domain containing 6 |
| chr15_-_67048595 | 0.15 |
ENSMUST00000229213.2
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr19_-_8700728 | 0.15 |
ENSMUST00000170157.8
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr17_+_28075495 | 0.15 |
ENSMUST00000233809.2
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr10_-_62285458 | 0.15 |
ENSMUST00000020273.16
|
Supv3l1
|
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr14_-_57635969 | 0.15 |
ENSMUST00000022517.9
|
Cryl1
|
crystallin, lambda 1 |
| chr4_+_34614939 | 0.15 |
ENSMUST00000029968.14
ENSMUST00000148519.3 |
Rars2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr12_-_4924341 | 0.15 |
ENSMUST00000137337.8
ENSMUST00000045921.14 |
Mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr5_+_114706077 | 0.15 |
ENSMUST00000043650.8
|
Fam222a
|
family with sequence similarity 222, member A |
| chr10_-_90959853 | 0.14 |
ENSMUST00000170810.8
ENSMUST00000076694.13 |
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr4_+_10874498 | 0.14 |
ENSMUST00000080517.14
|
2610301B20Rik
|
RIKEN cDNA 2610301B20 gene |
| chr15_-_83479312 | 0.14 |
ENSMUST00000016901.5
|
Ttll12
|
tubulin tyrosine ligase-like family, member 12 |
| chr2_-_109111064 | 0.14 |
ENSMUST00000147770.2
|
Mettl15
|
methyltransferase like 15 |
| chr5_-_31350352 | 0.14 |
ENSMUST00000202758.4
ENSMUST00000114603.8 |
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr12_+_111504450 | 0.13 |
ENSMUST00000166123.9
ENSMUST00000222441.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr1_+_139349912 | 0.13 |
ENSMUST00000200243.5
ENSMUST00000039867.10 |
Zbtb41
|
zinc finger and BTB domain containing 41 |
| chr3_+_41697046 | 0.13 |
ENSMUST00000120167.8
ENSMUST00000108065.9 ENSMUST00000146165.8 ENSMUST00000192193.6 ENSMUST00000119572.8 ENSMUST00000026867.14 ENSMUST00000026868.13 |
D3Ertd751e
|
DNA segment, Chr 3, ERATO Doi 751, expressed |
| chr19_-_4319170 | 0.13 |
ENSMUST00000037992.16
ENSMUST00000113852.6 ENSMUST00000236794.2 |
Ssh3
|
slingshot protein phosphatase 3 |
| chr5_-_148336711 | 0.13 |
ENSMUST00000048116.15
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr7_-_121580930 | 0.13 |
ENSMUST00000205438.2
ENSMUST00000057576.8 |
Cog7
|
component of oligomeric golgi complex 7 |
| chr18_+_11790888 | 0.13 |
ENSMUST00000234499.2
|
Rbbp8
|
retinoblastoma binding protein 8, endonuclease |
| chr3_-_68911886 | 0.13 |
ENSMUST00000169064.8
|
Ift80
|
intraflagellar transport 80 |
| chr3_-_51248032 | 0.12 |
ENSMUST00000062009.14
ENSMUST00000194641.6 |
Elf2
|
E74-like factor 2 |
| chr5_-_65855199 | 0.12 |
ENSMUST00000031104.7
|
Pds5a
|
PDS5 cohesin associated factor A |
| chr13_+_84370405 | 0.12 |
ENSMUST00000057495.10
ENSMUST00000225069.2 |
Tmem161b
|
transmembrane protein 161B |
| chr6_+_71808427 | 0.11 |
ENSMUST00000171057.2
|
Immt
|
inner membrane protein, mitochondrial |
| chr16_+_12927547 | 0.11 |
ENSMUST00000023206.14
|
Ercc4
|
excision repair cross-complementing rodent repair deficiency, complementation group 4 |
| chr8_+_123294740 | 0.10 |
ENSMUST00000006760.3
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr2_-_104542467 | 0.10 |
ENSMUST00000111118.8
ENSMUST00000028597.4 |
Tcp11l1
|
t-complex 11 like 1 |
| chr12_+_8971603 | 0.10 |
ENSMUST00000020909.4
|
Laptm4a
|
lysosomal-associated protein transmembrane 4A |
| chr16_+_90017634 | 0.10 |
ENSMUST00000023707.11
|
Sod1
|
superoxide dismutase 1, soluble |
| chr4_+_3938903 | 0.10 |
ENSMUST00000121210.8
ENSMUST00000121651.8 ENSMUST00000041122.11 ENSMUST00000120732.8 ENSMUST00000119307.8 ENSMUST00000123769.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_-_34614883 | 0.10 |
ENSMUST00000140334.2
ENSMUST00000108142.8 ENSMUST00000048706.10 |
Orc3
|
origin recognition complex, subunit 3 |
| chr11_+_84020475 | 0.10 |
ENSMUST00000133811.3
|
Acaca
|
acetyl-Coenzyme A carboxylase alpha |
| chr11_+_60066681 | 0.09 |
ENSMUST00000102688.2
|
Rai1
|
retinoic acid induced 1 |
| chr16_-_78173645 | 0.09 |
ENSMUST00000023570.14
|
Btg3
|
BTG anti-proliferation factor 3 |
| chr8_+_71995554 | 0.09 |
ENSMUST00000034272.9
|
Mvb12a
|
multivesicular body subunit 12A |
| chr9_-_78350486 | 0.09 |
ENSMUST00000070742.14
ENSMUST00000034898.14 |
Cgas
|
cyclic GMP-AMP synthase |
| chr19_+_38384428 | 0.09 |
ENSMUST00000054098.4
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr4_+_11486002 | 0.09 |
ENSMUST00000108307.3
|
Virma
|
vir like m6A methyltransferase associated |
| chr5_+_117457126 | 0.09 |
ENSMUST00000111967.8
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr2_+_180240015 | 0.09 |
ENSMUST00000103059.2
|
Col9a3
|
collagen, type IX, alpha 3 |
| chr5_-_148336574 | 0.09 |
ENSMUST00000202457.4
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr12_+_111505216 | 0.09 |
ENSMUST00000050993.11
|
Eif5
|
eukaryotic translation initiation factor 5 |
| chr1_+_90531183 | 0.09 |
ENSMUST00000186750.2
|
Cops8
|
COP9 signalosome subunit 8 |
| chr13_-_92491451 | 0.09 |
ENSMUST00000187424.2
ENSMUST00000185852.7 ENSMUST00000022220.13 |
Msh3
|
mutS homolog 3 |
| chr13_-_55477535 | 0.08 |
ENSMUST00000021941.8
|
Mxd3
|
Max dimerization protein 3 |
| chr3_-_68911835 | 0.08 |
ENSMUST00000107812.8
|
Ift80
|
intraflagellar transport 80 |
| chrX_-_50294652 | 0.08 |
ENSMUST00000114875.8
|
Mbnl3
|
muscleblind like splicing factor 3 |
| chr3_+_33854305 | 0.08 |
ENSMUST00000196139.5
ENSMUST00000200271.5 ENSMUST00000198529.5 ENSMUST00000117915.8 ENSMUST00000108210.9 ENSMUST00000196975.5 |
Ttc14
|
tetratricopeptide repeat domain 14 |
| chrX_-_104973003 | 0.08 |
ENSMUST00000130980.2
ENSMUST00000113573.8 |
Atrx
|
ATRX, chromatin remodeler |
| chr7_+_23781311 | 0.08 |
ENSMUST00000207002.2
ENSMUST00000068975.6 ENSMUST00000203854.3 |
Zfp180
|
zinc finger protein 180 |
| chr12_-_108145498 | 0.08 |
ENSMUST00000071095.14
|
Setd3
|
SET domain containing 3 |
| chr2_+_30282414 | 0.08 |
ENSMUST00000123202.8
ENSMUST00000113612.10 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr6_+_47431254 | 0.08 |
ENSMUST00000031697.9
ENSMUST00000143941.3 |
Cul1
|
cullin 1 |
| chr2_+_152804405 | 0.08 |
ENSMUST00000099197.9
ENSMUST00000103155.10 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr18_+_44513547 | 0.08 |
ENSMUST00000202306.2
ENSMUST00000025350.10 |
Dcp2
|
decapping mRNA 2 |
| chr6_+_71808309 | 0.08 |
ENSMUST00000064062.13
ENSMUST00000166975.8 ENSMUST00000165331.8 ENSMUST00000114151.10 ENSMUST00000101301.10 ENSMUST00000166938.8 |
Immt
|
inner membrane protein, mitochondrial |
| chr11_+_101443014 | 0.07 |
ENSMUST00000147239.8
|
Nbr1
|
NBR1, autophagy cargo receptor |
| chr2_+_24944407 | 0.07 |
ENSMUST00000102931.11
ENSMUST00000074422.14 ENSMUST00000132172.8 ENSMUST00000114388.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr3_+_66893031 | 0.07 |
ENSMUST00000046542.13
ENSMUST00000162693.8 |
Rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr10_+_43355113 | 0.06 |
ENSMUST00000040147.8
|
Bend3
|
BEN domain containing 3 |
| chr2_-_180929828 | 0.06 |
ENSMUST00000049032.13
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr3_+_103483397 | 0.06 |
ENSMUST00000183637.8
ENSMUST00000117221.9 ENSMUST00000118117.8 ENSMUST00000118563.3 |
Syt6
|
synaptotagmin VI |
| chr9_-_105372330 | 0.06 |
ENSMUST00000038118.15
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr7_-_5016237 | 0.06 |
ENSMUST00000208944.2
|
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr18_+_9957906 | 0.06 |
ENSMUST00000025137.9
|
Thoc1
|
THO complex 1 |
| chr12_+_111504640 | 0.06 |
ENSMUST00000222375.2
ENSMUST00000222388.2 |
Eif5
|
eukaryotic translation initiation factor 5 |
| chr7_-_100661220 | 0.05 |
ENSMUST00000207916.2
|
P2ry2
|
purinergic receptor P2Y, G-protein coupled 2 |
| chr7_-_30643444 | 0.05 |
ENSMUST00000062620.9
|
Hamp
|
hepcidin antimicrobial peptide |
| chr10_+_43354807 | 0.05 |
ENSMUST00000167488.9
|
Bend3
|
BEN domain containing 3 |
| chr18_-_52662728 | 0.04 |
ENSMUST00000025409.9
|
Lox
|
lysyl oxidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.2 | 1.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 0.5 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 0.2 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.2 | 0.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 3.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.5 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.5 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.1 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.4 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.3 | GO:0090650 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.2 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.4 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.7 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 0.3 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.2 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.1 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.1 | 0.4 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.2 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.2 | GO:0046724 | oxalic acid secretion(GO:0046724) |
| 0.1 | 0.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.1 | 0.7 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.4 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.4 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0051309 | female meiosis chromosome separation(GO:0051309) |
| 0.0 | 0.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.7 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.7 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 0.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.4 | GO:0097632 | extrinsic component of pre-autophagosomal structure membrane(GO:0097632) |
| 0.1 | 0.3 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 0.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.4 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.5 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.4 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 1.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.7 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.6 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0032142 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 1.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0047874 | dolichyldiphosphatase activity(GO:0047874) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.7 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 2.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.7 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |