Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
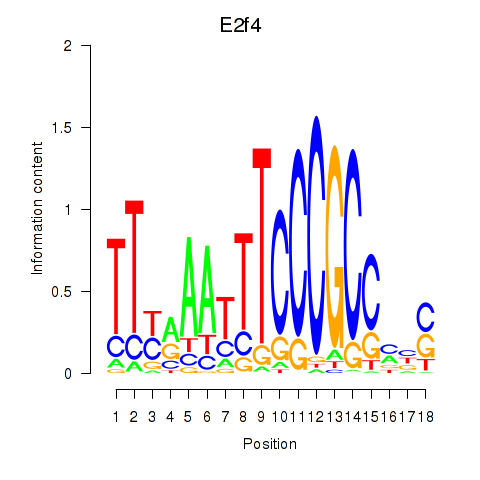
Results for E2f4
Z-value: 0.67
Transcription factors associated with E2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f4
|
ENSMUSG00000014859.10 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f4 | mm39_v1_chr8_+_106024294_106024367 | -0.96 | 8.1e-03 | Click! |
Activity profile of E2f4 motif
Sorted Z-values of E2f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_133694607 | 0.49 |
ENSMUST00000105893.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr12_+_24758240 | 0.41 |
ENSMUST00000020980.12
|
Rrm2
|
ribonucleotide reductase M2 |
| chr4_+_134238310 | 0.40 |
ENSMUST00000105866.3
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr6_-_56681657 | 0.35 |
ENSMUST00000176595.3
ENSMUST00000170382.5 ENSMUST00000203958.2 |
Lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chrX_+_37689503 | 0.31 |
ENSMUST00000000365.3
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr1_-_72251466 | 0.31 |
ENSMUST00000048860.9
|
Mreg
|
melanoregulin |
| chr1_-_10108325 | 0.31 |
ENSMUST00000027050.10
ENSMUST00000188619.2 |
Cops5
|
COP9 signalosome subunit 5 |
| chr10_+_87982916 | 0.29 |
ENSMUST00000169309.3
|
Nup37
|
nucleoporin 37 |
| chr5_+_21577640 | 0.29 |
ENSMUST00000035799.6
|
Fgl2
|
fibrinogen-like protein 2 |
| chr10_+_87982854 | 0.28 |
ENSMUST00000052355.15
|
Nup37
|
nucleoporin 37 |
| chr17_+_85264134 | 0.28 |
ENSMUST00000112305.10
|
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr4_-_133694543 | 0.28 |
ENSMUST00000123234.8
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_68936457 | 0.28 |
ENSMUST00000108666.8
ENSMUST00000021277.6 |
Aurkb
|
aurora kinase B |
| chr4_-_132072988 | 0.27 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr8_+_108271643 | 0.26 |
ENSMUST00000212543.2
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_57416752 | 0.26 |
ENSMUST00000042734.3
|
1700066M21Rik
|
RIKEN cDNA 1700066M21 gene |
| chr3_-_107876477 | 0.26 |
ENSMUST00000004136.10
|
Gstm3
|
glutathione S-transferase, mu 3 |
| chr4_-_132073048 | 0.26 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr9_+_107828136 | 0.25 |
ENSMUST00000049348.9
ENSMUST00000194271.2 |
Traip
|
TRAF-interacting protein |
| chr11_+_95557783 | 0.24 |
ENSMUST00000125172.8
ENSMUST00000036374.6 |
Phb
|
prohibitin |
| chr19_+_6135013 | 0.24 |
ENSMUST00000025704.3
|
Cdca5
|
cell division cycle associated 5 |
| chr4_-_133695264 | 0.24 |
ENSMUST00000102553.11
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_-_133695204 | 0.24 |
ENSMUST00000100472.10
ENSMUST00000136327.2 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chrX_+_12454031 | 0.23 |
ENSMUST00000033313.3
|
Atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr17_-_66408461 | 0.22 |
ENSMUST00000024909.15
ENSMUST00000147484.2 |
Ndufv2
|
NADH:ubiquinone oxidoreductase core subunit V2 |
| chr1_-_169359015 | 0.22 |
ENSMUST00000111368.8
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr4_-_3835595 | 0.22 |
ENSMUST00000138502.2
|
Rps20
|
ribosomal protein S20 |
| chr9_+_106354463 | 0.21 |
ENSMUST00000047721.10
|
Rrp9
|
ribosomal RNA processing 9, U3 small nucleolar RNA binding protein |
| chr2_+_125994050 | 0.21 |
ENSMUST00000170908.8
|
Dtwd1
|
DTW domain containing 1 |
| chr19_-_7460550 | 0.20 |
ENSMUST00000088169.7
|
Rtn3
|
reticulon 3 |
| chr5_-_138170077 | 0.20 |
ENSMUST00000155902.8
ENSMUST00000148879.8 |
Mcm7
|
minichromosome maintenance complex component 7 |
| chr2_-_3513783 | 0.20 |
ENSMUST00000124331.8
ENSMUST00000140494.2 ENSMUST00000027961.12 |
Gm45902
Hspa14
|
predicted gene 45902 heat shock protein 14 |
| chr5_-_92496730 | 0.20 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr15_-_98729333 | 0.20 |
ENSMUST00000168846.3
|
Prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr12_-_87194658 | 0.19 |
ENSMUST00000037788.6
|
Pomt2
|
protein-O-mannosyltransferase 2 |
| chrX_+_163763588 | 0.18 |
ENSMUST00000167446.8
ENSMUST00000057150.8 |
Fancb
|
Fanconi anemia, complementation group B |
| chr11_+_108811168 | 0.18 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr11_+_108316018 | 0.18 |
ENSMUST00000150863.9
ENSMUST00000061287.12 ENSMUST00000149683.9 |
Cep112
|
centrosomal protein 112 |
| chr9_-_103243039 | 0.17 |
ENSMUST00000035484.11
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr2_+_71617402 | 0.17 |
ENSMUST00000238991.2
|
Itga6
|
integrin alpha 6 |
| chr2_+_71617266 | 0.17 |
ENSMUST00000112101.8
ENSMUST00000028522.10 |
Itga6
|
integrin alpha 6 |
| chr9_+_120762884 | 0.16 |
ENSMUST00000145093.2
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr11_+_120675083 | 0.16 |
ENSMUST00000100134.10
ENSMUST00000208737.2 |
Gps1
|
G protein pathway suppressor 1 |
| chr11_-_51579441 | 0.16 |
ENSMUST00000007921.9
|
0610009B22Rik
|
RIKEN cDNA 0610009B22 gene |
| chr11_-_5492175 | 0.16 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr17_-_25946370 | 0.16 |
ENSMUST00000170070.3
ENSMUST00000048054.14 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr1_+_85822250 | 0.15 |
ENSMUST00000185569.2
|
Itm2c
|
integral membrane protein 2C |
| chr1_-_169358912 | 0.15 |
ENSMUST00000192248.2
ENSMUST00000028000.13 |
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr9_-_82857528 | 0.15 |
ENSMUST00000034787.12
|
Phip
|
pleckstrin homology domain interacting protein |
| chr19_-_7460600 | 0.15 |
ENSMUST00000235593.2
ENSMUST00000088171.12 ENSMUST00000065304.13 ENSMUST00000025667.14 |
Rtn3
|
reticulon 3 |
| chrX_+_161684221 | 0.14 |
ENSMUST00000101095.9
|
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr18_-_34884555 | 0.14 |
ENSMUST00000060710.9
|
Cdc25c
|
cell division cycle 25C |
| chr4_+_135583018 | 0.14 |
ENSMUST00000105853.10
ENSMUST00000097844.9 ENSMUST00000102544.9 ENSMUST00000126641.2 |
Srsf10
|
serine and arginine-rich splicing factor 10 |
| chr8_+_124750133 | 0.14 |
ENSMUST00000034457.9
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr10_-_87982732 | 0.14 |
ENSMUST00000164121.8
ENSMUST00000164803.2 ENSMUST00000168163.8 ENSMUST00000048518.16 |
Parpbp
|
PARP1 binding protein |
| chrX_-_163763273 | 0.14 |
ENSMUST00000112247.9
ENSMUST00000004715.2 |
Mospd2
|
motile sperm domain containing 2 |
| chr14_+_76725876 | 0.13 |
ENSMUST00000101618.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr6_+_124807176 | 0.13 |
ENSMUST00000131847.8
ENSMUST00000151674.3 |
Cdca3
|
cell division cycle associated 3 |
| chr17_-_35265702 | 0.13 |
ENSMUST00000097338.11
|
Msh5
|
mutS homolog 5 |
| chrX_+_141464393 | 0.13 |
ENSMUST00000112889.8
ENSMUST00000101198.9 ENSMUST00000112891.8 ENSMUST00000087333.9 |
Tmem164
|
transmembrane protein 164 |
| chr5_+_130285870 | 0.13 |
ENSMUST00000100662.10
ENSMUST00000040213.13 |
Tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr3_-_62414391 | 0.13 |
ENSMUST00000029336.6
|
Dhx36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr4_+_44004437 | 0.12 |
ENSMUST00000107846.10
|
Clta
|
clathrin, light polypeptide (Lca) |
| chr10_+_36850532 | 0.12 |
ENSMUST00000019911.14
ENSMUST00000105510.2 |
Hdac2
|
histone deacetylase 2 |
| chr1_+_9868332 | 0.12 |
ENSMUST00000166384.8
ENSMUST00000168907.8 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr12_+_105976463 | 0.12 |
ENSMUST00000072040.7
|
Vrk1
|
vaccinia related kinase 1 |
| chr15_-_10714653 | 0.12 |
ENSMUST00000169385.3
|
Rai14
|
retinoic acid induced 14 |
| chr11_-_106890307 | 0.12 |
ENSMUST00000018506.13
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr14_+_55815580 | 0.11 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr12_+_117807607 | 0.11 |
ENSMUST00000176735.8
ENSMUST00000177339.2 |
Cdca7l
|
cell division cycle associated 7 like |
| chr15_-_39807359 | 0.11 |
ENSMUST00000110305.3
|
Lrp12
|
low density lipoprotein-related protein 12 |
| chr12_-_118162652 | 0.11 |
ENSMUST00000084806.7
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chr2_+_162896602 | 0.11 |
ENSMUST00000018005.10
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr4_-_115980813 | 0.11 |
ENSMUST00000102704.4
ENSMUST00000102705.10 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr19_+_8713156 | 0.11 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr19_+_5416769 | 0.11 |
ENSMUST00000025759.9
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr1_+_92862432 | 0.11 |
ENSMUST00000117814.8
|
Capn10
|
calpain 10 |
| chr18_+_65933586 | 0.10 |
ENSMUST00000025394.14
ENSMUST00000236847.2 ENSMUST00000153193.3 |
Sec11c
|
SEC11 homolog C, signal peptidase complex subunit |
| chr8_-_11600689 | 0.10 |
ENSMUST00000049461.7
|
Cars2
|
cysteinyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr18_+_56840813 | 0.10 |
ENSMUST00000025486.9
|
Lmnb1
|
lamin B1 |
| chr19_+_45991907 | 0.10 |
ENSMUST00000099393.4
|
Hps6
|
HPS6, biogenesis of lysosomal organelles complex 2 subunit 3 |
| chrX_-_165992145 | 0.10 |
ENSMUST00000112176.8
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr19_+_6451667 | 0.10 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr17_+_35219998 | 0.10 |
ENSMUST00000173584.8
|
Vars
|
valyl-tRNA synthetase |
| chr7_-_37806912 | 0.10 |
ENSMUST00000108023.10
|
Ccne1
|
cyclin E1 |
| chr3_-_142587419 | 0.09 |
ENSMUST00000174422.8
ENSMUST00000173830.8 |
Pkn2
|
protein kinase N2 |
| chr4_+_108691798 | 0.09 |
ENSMUST00000030296.9
|
Txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr5_-_138170644 | 0.09 |
ENSMUST00000000505.16
|
Mcm7
|
minichromosome maintenance complex component 7 |
| chr2_+_180961507 | 0.09 |
ENSMUST00000098971.11
ENSMUST00000054622.15 ENSMUST00000108814.8 ENSMUST00000048608.16 ENSMUST00000108815.8 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr5_+_123214332 | 0.09 |
ENSMUST00000067505.15
ENSMUST00000111619.10 ENSMUST00000160344.2 |
Tmem120b
|
transmembrane protein 120B |
| chr4_-_59783780 | 0.09 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr9_+_44245981 | 0.08 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr13_-_74085880 | 0.08 |
ENSMUST00000022053.11
|
Trip13
|
thyroid hormone receptor interactor 13 |
| chr5_-_25910788 | 0.08 |
ENSMUST00000030773.12
|
Xrcc2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr8_-_106223502 | 0.08 |
ENSMUST00000212303.2
|
Zdhhc1
|
zinc finger, DHHC domain containing 1 |
| chr16_-_59452883 | 0.08 |
ENSMUST00000118438.8
|
Arl6
|
ADP-ribosylation factor-like 6 |
| chr11_+_77928736 | 0.08 |
ENSMUST00000072289.12
ENSMUST00000100784.9 ENSMUST00000073660.7 |
Flot2
|
flotillin 2 |
| chr12_+_105976533 | 0.07 |
ENSMUST00000085026.12
ENSMUST00000021539.16 |
Vrk1
|
vaccinia related kinase 1 |
| chr9_+_57615816 | 0.07 |
ENSMUST00000043990.14
ENSMUST00000142807.2 |
Edc3
|
enhancer of mRNA decapping 3 |
| chr17_+_35219941 | 0.07 |
ENSMUST00000087315.14
|
Vars
|
valyl-tRNA synthetase |
| chr3_+_88461059 | 0.07 |
ENSMUST00000008748.8
|
Ubqln4
|
ubiquilin 4 |
| chr1_-_157240138 | 0.07 |
ENSMUST00000078308.13
|
Rasal2
|
RAS protein activator like 2 |
| chr9_-_60595401 | 0.07 |
ENSMUST00000114034.9
ENSMUST00000065603.12 |
Lrrc49
|
leucine rich repeat containing 49 |
| chr17_-_66408543 | 0.07 |
ENSMUST00000143987.9
|
Ndufv2
|
NADH:ubiquinone oxidoreductase core subunit V2 |
| chr3_-_116456292 | 0.07 |
ENSMUST00000029570.9
|
Mfsd14a
|
major facilitator superfamily domain containing 14A |
| chr11_-_101442663 | 0.07 |
ENSMUST00000017290.11
|
Brca1
|
breast cancer 1, early onset |
| chr11_+_101443014 | 0.07 |
ENSMUST00000147239.8
|
Nbr1
|
NBR1, autophagy cargo receptor |
| chr9_-_103242737 | 0.06 |
ENSMUST00000072249.13
ENSMUST00000189896.2 |
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr11_+_29080733 | 0.06 |
ENSMUST00000020756.9
|
Pnpt1
|
polyribonucleotide nucleotidyltransferase 1 |
| chrX_+_104123341 | 0.06 |
ENSMUST00000033577.11
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr5_+_138170259 | 0.06 |
ENSMUST00000019662.11
ENSMUST00000151318.8 |
Ap4m1
|
adaptor-related protein complex AP-4, mu 1 |
| chr10_-_90959853 | 0.06 |
ENSMUST00000170810.8
ENSMUST00000076694.13 |
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chrX_+_161684563 | 0.06 |
ENSMUST00000112303.8
ENSMUST00000033727.14 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr9_-_83323080 | 0.06 |
ENSMUST00000034791.15
ENSMUST00000188548.7 ENSMUST00000034793.15 |
Lca5
|
Leber congenital amaurosis 5 (human) |
| chr8_-_54091980 | 0.06 |
ENSMUST00000047768.11
|
Neil3
|
nei like 3 (E. coli) |
| chr2_-_145776934 | 0.06 |
ENSMUST00000001818.5
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr6_-_113508536 | 0.05 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr7_+_126808016 | 0.05 |
ENSMUST00000206204.2
ENSMUST00000206772.2 |
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr3_+_89680867 | 0.05 |
ENSMUST00000038356.13
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chrX_+_104123367 | 0.05 |
ENSMUST00000119477.2
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr3_-_142587678 | 0.05 |
ENSMUST00000043812.15
|
Pkn2
|
protein kinase N2 |
| chrX_+_161684735 | 0.05 |
ENSMUST00000112302.8
ENSMUST00000112301.8 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr1_-_179631190 | 0.05 |
ENSMUST00000027768.14
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr2_+_180961599 | 0.05 |
ENSMUST00000153112.2
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr7_+_24584076 | 0.04 |
ENSMUST00000153451.9
ENSMUST00000108429.8 |
Rps19
|
ribosomal protein S19 |
| chr11_+_108811626 | 0.04 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr2_-_52566583 | 0.04 |
ENSMUST00000178799.8
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr17_+_31514780 | 0.04 |
ENSMUST00000237460.2
|
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr5_-_33809640 | 0.04 |
ENSMUST00000151081.2
ENSMUST00000139518.8 ENSMUST00000101354.10 |
Slbp
|
stem-loop binding protein |
| chr17_+_31515163 | 0.03 |
ENSMUST00000235972.2
ENSMUST00000165149.3 ENSMUST00000236251.2 |
Slc37a1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1 |
| chr12_+_99850756 | 0.03 |
ENSMUST00000153627.8
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr2_-_30176324 | 0.03 |
ENSMUST00000100219.5
|
Dolk
|
dolichol kinase |
| chr11_-_106890195 | 0.03 |
ENSMUST00000106768.2
ENSMUST00000144834.8 |
Kpna2
|
karyopherin (importin) alpha 2 |
| chr13_-_14237958 | 0.03 |
ENSMUST00000223174.2
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr17_+_25946644 | 0.03 |
ENSMUST00000237183.2
ENSMUST00000237785.2 ENSMUST00000047273.3 |
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr2_-_62476487 | 0.03 |
ENSMUST00000112459.4
ENSMUST00000028259.12 |
Ifih1
|
interferon induced with helicase C domain 1 |
| chr5_-_33809872 | 0.03 |
ENSMUST00000057551.14
|
Slbp
|
stem-loop binding protein |
| chrX_-_163763337 | 0.02 |
ENSMUST00000112248.9
|
Mospd2
|
motile sperm domain containing 2 |
| chr4_+_43059028 | 0.02 |
ENSMUST00000163653.8
ENSMUST00000107952.9 ENSMUST00000107953.9 |
Unc13b
|
unc-13 homolog B |
| chr7_-_97228589 | 0.02 |
ENSMUST00000151840.2
ENSMUST00000135998.8 ENSMUST00000144858.8 ENSMUST00000146605.8 ENSMUST00000072725.12 ENSMUST00000138060.3 ENSMUST00000154853.8 ENSMUST00000136757.8 ENSMUST00000124552.3 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr12_-_4283926 | 0.02 |
ENSMUST00000111169.10
ENSMUST00000020981.12 |
Cenpo
|
centromere protein O |
| chr16_-_90524214 | 0.02 |
ENSMUST00000099554.5
|
Mis18a
|
MIS18 kinetochore protein A |
| chr7_-_92523396 | 0.02 |
ENSMUST00000209074.2
ENSMUST00000208356.2 ENSMUST00000032877.11 |
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr14_-_54754810 | 0.02 |
ENSMUST00000023873.12
|
Prmt5
|
protein arginine N-methyltransferase 5 |
| chr6_-_148732893 | 0.02 |
ENSMUST00000145960.2
|
Ipo8
|
importin 8 |
| chr4_+_11558905 | 0.02 |
ENSMUST00000095145.12
ENSMUST00000108306.9 ENSMUST00000070755.13 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr7_-_81104423 | 0.02 |
ENSMUST00000178892.3
ENSMUST00000098331.10 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr19_+_4771089 | 0.02 |
ENSMUST00000238976.3
|
Sptbn2
|
spectrin beta, non-erythrocytic 2 |
| chr6_+_113508636 | 0.02 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr2_+_24257576 | 0.02 |
ENSMUST00000140547.2
ENSMUST00000102942.8 |
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr15_-_37792635 | 0.01 |
ENSMUST00000090150.11
ENSMUST00000150453.2 |
Ncald
|
neurocalcin delta |
| chr9_-_123507847 | 0.01 |
ENSMUST00000170591.2
ENSMUST00000171647.9 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr7_+_45433103 | 0.01 |
ENSMUST00000209617.2
ENSMUST00000209701.2 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr15_-_37792237 | 0.01 |
ENSMUST00000168992.8
ENSMUST00000148652.9 |
Ncald
|
neurocalcin delta |
| chr11_+_74510413 | 0.01 |
ENSMUST00000100866.3
|
Ccdc92b
|
coiled-coil domain containing 92B |
| chr7_+_97020810 | 0.01 |
ENSMUST00000098300.6
|
Alg8
|
asparagine-linked glycosylation 8 (alpha-1,3-glucosyltransferase) |
| chr7_-_126859790 | 0.01 |
ENSMUST00000035276.5
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr3_+_87704258 | 0.01 |
ENSMUST00000029711.9
ENSMUST00000107582.3 |
Insrr
|
insulin receptor-related receptor |
| chr17_+_35220252 | 0.01 |
ENSMUST00000174260.8
|
Vars
|
valyl-tRNA synthetase |
| chrY_-_90850446 | 0.01 |
ENSMUST00000179623.2
|
Gm21748
|
predicted gene, 21748 |
| chr2_-_63014514 | 0.01 |
ENSMUST00000112452.2
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr14_+_70793340 | 0.01 |
ENSMUST00000163060.2
|
Hr
|
lysine demethylase and nuclear receptor corepressor |
| chr1_+_52669849 | 0.01 |
ENSMUST00000165859.8
|
Nemp2
|
nuclear envelope integral membrane protein 2 |
| chr2_+_101508747 | 0.01 |
ENSMUST00000004949.8
|
Traf6
|
TNF receptor-associated factor 6 |
| chr17_+_88748139 | 0.01 |
ENSMUST00000112238.9
ENSMUST00000155640.2 |
Foxn2
|
forkhead box N2 |
| chr13_-_41264511 | 0.01 |
ENSMUST00000225271.2
|
Gcm2
|
glial cells missing homolog 2 |
| chr5_+_30437579 | 0.01 |
ENSMUST00000145167.9
|
Selenoi
|
selenoprotein I |
| chr2_+_106525938 | 0.01 |
ENSMUST00000016530.14
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr12_-_104120105 | 0.01 |
ENSMUST00000085050.4
|
Serpina3c
|
serine (or cysteine) peptidase inhibitor, clade A, member 3C |
| chr7_+_45433306 | 0.01 |
ENSMUST00000072580.12
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr18_-_58343423 | 0.01 |
ENSMUST00000025497.8
|
Fbn2
|
fibrillin 2 |
| chr11_+_16901370 | 0.01 |
ENSMUST00000109635.2
ENSMUST00000061327.2 |
Fbxo48
|
F-box protein 48 |
| chr19_+_46064302 | 0.01 |
ENSMUST00000165017.2
ENSMUST00000223741.2 ENSMUST00000225780.2 |
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr3_-_6685492 | 0.01 |
ENSMUST00000091364.4
|
1700008P02Rik
|
RIKEN cDNA 1700008P02 gene |
| chr4_+_43562706 | 0.00 |
ENSMUST00000167751.2
ENSMUST00000132631.2 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr8_-_123884968 | 0.00 |
ENSMUST00000137998.2
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr5_-_108697857 | 0.00 |
ENSMUST00000129040.2
ENSMUST00000046892.10 |
Cplx1
|
complexin 1 |
| chr4_-_44704006 | 0.00 |
ENSMUST00000146335.8
|
Pax5
|
paired box 5 |
| chr4_-_58553171 | 0.00 |
ENSMUST00000145361.2
|
Lpar1
|
lysophosphatidic acid receptor 1 |
| chr8_-_107783282 | 0.00 |
ENSMUST00000034391.4
ENSMUST00000095517.12 |
Cog8
|
component of oligomeric golgi complex 8 |
| chr14_+_33073249 | 0.00 |
ENSMUST00000140711.3
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr10_+_12936248 | 0.00 |
ENSMUST00000193426.6
|
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr2_+_163046394 | 0.00 |
ENSMUST00000128999.8
|
Tox2
|
TOX high mobility group box family member 2 |
| chr1_-_33708876 | 0.00 |
ENSMUST00000027312.11
|
Prim2
|
DNA primase, p58 subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 0.2 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.2 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 0.0 | 0.2 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 1.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.3 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.2 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:1904008 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.9 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.3 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 0.2 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:1903903 | protein localization to plasma membrane raft(GO:0044860) regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.3 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.0 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:0003352 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.2 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |