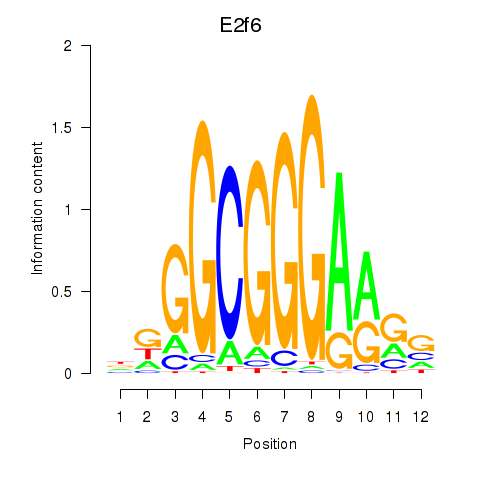
|
chr13_-_21934675
Show fit
|
0.71 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12
|
|
chr13_-_23735822
Show fit
|
0.60 |
ENSMUST00000102971.2
|
H4c6
|
H4 clustered histone 6
|
|
chr3_-_96170627
Show fit
|
0.59 |
ENSMUST00000171473.3
|
H4c14
|
H4 clustered histone 14
|
|
chr6_+_42326714
Show fit
|
0.55 |
ENSMUST00000203846.3
|
Zyx
|
zyxin
|
|
chr1_-_71692320
Show fit
|
0.50 |
ENSMUST00000186940.7
ENSMUST00000188894.7
ENSMUST00000188674.7
ENSMUST00000189821.7
ENSMUST00000187938.7
ENSMUST00000190780.7
ENSMUST00000186736.2
ENSMUST00000055226.13
ENSMUST00000186129.7
|
Fn1
|
fibronectin 1
|
|
chr13_+_23765546
Show fit
|
0.48 |
ENSMUST00000102968.3
|
H4c4
|
H4 clustered histone 4
|
|
chr6_+_42326980
Show fit
|
0.42 |
ENSMUST00000203849.2
|
Zyx
|
zyxin
|
|
chr16_+_43993599
Show fit
|
0.38 |
ENSMUST00000119746.8
ENSMUST00000088356.10
ENSMUST00000169582.3
|
Usf3
|
upstream transcription factor family member 3
|
|
chr6_+_42326934
Show fit
|
0.38 |
ENSMUST00000203401.3
ENSMUST00000164375.4
|
Zyx
|
zyxin
|
|
chr7_-_127423641
Show fit
|
0.35 |
ENSMUST00000106267.5
|
Stx1b
|
syntaxin 1B
|
|
chr7_-_100581314
Show fit
|
0.35 |
ENSMUST00000107032.3
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17
|
|
chr2_-_155315708
Show fit
|
0.35 |
ENSMUST00000109670.8
ENSMUST00000123293.8
|
Ncoa6
|
nuclear receptor coactivator 6
|
|
chr6_+_42326760
Show fit
|
0.34 |
ENSMUST00000203652.3
ENSMUST00000070635.13
|
Zyx
|
zyxin
|
|
chr15_-_82783978
Show fit
|
0.32 |
ENSMUST00000230403.2
|
Tcf20
|
transcription factor 20
|
|
chr3_+_96177010
Show fit
|
0.31 |
ENSMUST00000051089.4
ENSMUST00000177113.2
|
Gm42743
H2bc18
|
predicted gene 42743
H2B clustered histone 18
|
|
chr11_-_5049082
Show fit
|
0.30 |
ENSMUST00000063232.7
|
Ewsr1
|
Ewing sarcoma breakpoint region 1
|
|
chr5_+_147366953
Show fit
|
0.29 |
ENSMUST00000031651.15
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit
|
|
chr8_-_85526653
Show fit
|
0.29 |
ENSMUST00000126806.2
ENSMUST00000076715.13
|
Nfix
|
nuclear factor I/X
|
|
chr5_-_84565218
Show fit
|
0.28 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5
|
|
chr8_-_85526972
Show fit
|
0.28 |
ENSMUST00000099070.10
|
Nfix
|
nuclear factor I/X
|
|
chr15_-_50753437
Show fit
|
0.28 |
ENSMUST00000077935.6
|
Trps1
|
transcriptional repressor GATA binding 1
|
|
chr11_-_96868483
Show fit
|
0.27 |
ENSMUST00000107624.8
|
Sp2
|
Sp2 transcription factor
|
|
chr7_+_100021425
Show fit
|
0.27 |
ENSMUST00000098259.11
ENSMUST00000051777.15
|
C2cd3
|
C2 calcium-dependent domain containing 3
|
|
chr5_+_138278777
Show fit
|
0.26 |
ENSMUST00000048028.15
ENSMUST00000162245.8
ENSMUST00000161691.2
|
Stag3
|
stromal antigen 3
|
|
chr15_-_66158445
Show fit
|
0.25 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3
|
|
chr9_-_50920236
Show fit
|
0.25 |
ENSMUST00000176824.8
ENSMUST00000176663.8
ENSMUST00000041375.11
|
Sik2
|
salt inducible kinase 2
|
|
chr1_-_74544946
Show fit
|
0.25 |
ENSMUST00000044260.11
ENSMUST00000186282.7
|
Usp37
|
ubiquitin specific peptidase 37
|
|
chr1_+_151446893
Show fit
|
0.25 |
ENSMUST00000134499.8
|
Niban1
|
niban apoptosis regulator 1
|
|
chr13_-_107158535
Show fit
|
0.25 |
ENSMUST00000117539.8
ENSMUST00000122233.8
ENSMUST00000022204.16
ENSMUST00000159772.8
|
Kif2a
|
kinesin family member 2A
|
|
chr4_-_141265873
Show fit
|
0.24 |
ENSMUST00000105786.3
|
Spen
|
spen family transcription repressor
|
|
chr5_-_76452577
Show fit
|
0.24 |
ENSMUST00000202651.4
|
Clock
|
circadian locomotor output cycles kaput
|
|
chr1_+_151447232
Show fit
|
0.24 |
ENSMUST00000111875.2
|
Niban1
|
niban apoptosis regulator 1
|
|
chr5_+_138278502
Show fit
|
0.23 |
ENSMUST00000160729.8
|
Stag3
|
stromal antigen 3
|
|
chr2_-_5719302
Show fit
|
0.23 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr4_-_141265745
Show fit
|
0.23 |
ENSMUST00000078886.10
|
Spen
|
spen family transcription repressor
|
|
chrX_-_7242065
Show fit
|
0.22 |
ENSMUST00000191497.2
ENSMUST00000115744.2
|
Usp27x
|
ubiquitin specific peptidase 27, X chromosome
|
|
chr18_+_7868823
Show fit
|
0.22 |
ENSMUST00000171042.8
ENSMUST00000166378.8
ENSMUST00000074919.11
|
Wac
|
WW domain containing adaptor with coiled-coil
|
|
chrX_-_72924436
Show fit
|
0.22 |
ENSMUST00000102871.10
|
L1cam
|
L1 cell adhesion molecule
|
|
chr15_+_79575046
Show fit
|
0.22 |
ENSMUST00000046463.10
|
Gtpbp1
|
GTP binding protein 1
|
|
chr12_+_76580386
Show fit
|
0.22 |
ENSMUST00000219063.2
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3
|
|
chr3_-_103698391
Show fit
|
0.22 |
ENSMUST00000106845.9
ENSMUST00000029438.15
|
Hipk1
|
homeodomain interacting protein kinase 1
|
|
chr5_+_137786031
Show fit
|
0.22 |
ENSMUST00000035852.14
|
Zcwpw1
|
zinc finger, CW type with PWWP domain 1
|
|
chr15_-_96540084
Show fit
|
0.21 |
ENSMUST00000230767.2
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr5_-_144294854
Show fit
|
0.21 |
ENSMUST00000055190.8
|
Baiap2l1
|
BAI1-associated protein 2-like 1
|
|
chr5_-_106844396
Show fit
|
0.21 |
ENSMUST00000137285.8
ENSMUST00000124263.2
ENSMUST00000112695.4
ENSMUST00000155495.8
ENSMUST00000135108.2
ENSMUST00000149128.3
|
Zfp644
Gm28039
|
zinc finger protein 644
predicted gene, 28039
|
|
chr1_+_151447124
Show fit
|
0.20 |
ENSMUST00000148810.8
|
Niban1
|
niban apoptosis regulator 1
|
|
chr12_+_84616536
Show fit
|
0.20 |
ENSMUST00000021665.12
ENSMUST00000169934.4
|
Vsx2
|
visual system homeobox 2
|
|
chr13_-_54835460
Show fit
|
0.20 |
ENSMUST00000129881.8
|
Rnf44
|
ring finger protein 44
|
|
chr18_+_7869066
Show fit
|
0.19 |
ENSMUST00000171486.8
ENSMUST00000170932.8
ENSMUST00000167020.8
|
Wac
|
WW domain containing adaptor with coiled-coil
|
|
chr7_-_29217967
Show fit
|
0.19 |
ENSMUST00000181975.8
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3
|
|
chr7_-_16348862
Show fit
|
0.19 |
ENSMUST00000171937.2
ENSMUST00000075845.11
|
Arhgap35
|
Rho GTPase activating protein 35
|
|
chr7_+_126380655
Show fit
|
0.19 |
ENSMUST00000172352.8
ENSMUST00000094037.5
|
Tbx6
|
T-box 6
|
|
chr12_-_76756772
Show fit
|
0.19 |
ENSMUST00000166101.2
|
Sptb
|
spectrin beta, erythrocytic
|
|
chr4_-_55532453
Show fit
|
0.19 |
ENSMUST00000132746.2
ENSMUST00000107619.3
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr5_-_90031180
Show fit
|
0.19 |
ENSMUST00000198151.2
|
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3
|
|
chr16_-_90081188
Show fit
|
0.19 |
ENSMUST00000163419.9
|
Scaf4
|
SR-related CTD-associated factor 4
|
|
chr15_-_76083575
Show fit
|
0.19 |
ENSMUST00000169438.8
|
Plec
|
plectin
|
|
chr19_+_23736205
Show fit
|
0.19 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1
|
|
chr15_-_58953838
Show fit
|
0.19 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1
|
|
chr2_-_160714473
Show fit
|
0.18 |
ENSMUST00000103111.9
|
Zhx3
|
zinc fingers and homeoboxes 3
|
|
chr17_-_81977590
Show fit
|
0.18 |
ENSMUST00000234923.2
|
Slc8a1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr18_-_84607615
Show fit
|
0.18 |
ENSMUST00000125763.3
|
Zfp407
|
zinc finger protein 407
|
|
chr19_+_10502679
Show fit
|
0.18 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7
|
|
chr18_+_31767369
Show fit
|
0.18 |
ENSMUST00000235017.2
ENSMUST00000178164.8
ENSMUST00000025109.8
|
Sap130
|
Sin3A associated protein
|
|
chr8_+_108669276
Show fit
|
0.18 |
ENSMUST00000220518.2
|
Zfhx3
|
zinc finger homeobox 3
|
|
chr8_+_105558204
Show fit
|
0.18 |
ENSMUST00000059449.7
|
Ces2b
|
carboxyesterase 2B
|
|
chr14_+_21550921
Show fit
|
0.18 |
ENSMUST00000182964.3
|
Kat6b
|
K(lysine) acetyltransferase 6B
|
|
chr1_-_36978602
Show fit
|
0.18 |
ENSMUST00000027290.12
|
Tmem131
|
transmembrane protein 131
|
|
chr17_+_36152383
Show fit
|
0.17 |
ENSMUST00000082337.13
|
Mdc1
|
mediator of DNA damage checkpoint 1
|
|
chr11_+_16702203
Show fit
|
0.17 |
ENSMUST00000102884.10
ENSMUST00000020329.13
|
Egfr
|
epidermal growth factor receptor
|
|
chr6_-_99498112
Show fit
|
0.17 |
ENSMUST00000177227.8
|
Foxp1
|
forkhead box P1
|
|
chr7_+_110367375
Show fit
|
0.17 |
ENSMUST00000170374.8
|
Ampd3
|
adenosine monophosphate deaminase 3
|
|
chr13_-_40887244
Show fit
|
0.17 |
ENSMUST00000110193.9
|
Tfap2a
|
transcription factor AP-2, alpha
|
|
chr11_+_49094292
Show fit
|
0.17 |
ENSMUST00000150284.8
ENSMUST00000109197.8
ENSMUST00000151228.2
|
Zfp62
|
zinc finger protein 62
|
|
chr4_-_123421441
Show fit
|
0.17 |
ENSMUST00000147228.8
|
Macf1
|
microtubule-actin crosslinking factor 1
|
|
chr7_+_44465714
Show fit
|
0.17 |
ENSMUST00000208172.2
|
Nup62
|
nucleoporin 62
|
|
chr19_+_53131187
Show fit
|
0.17 |
ENSMUST00000050096.15
ENSMUST00000237832.2
|
Add3
|
adducin 3 (gamma)
|
|
chr1_+_34044940
Show fit
|
0.17 |
ENSMUST00000187486.7
ENSMUST00000182697.8
|
Dst
|
dystonin
|
|
chr7_+_3648264
Show fit
|
0.16 |
ENSMUST00000206287.2
ENSMUST00000038913.16
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr9_+_100525807
Show fit
|
0.16 |
ENSMUST00000133388.2
|
Stag1
|
stromal antigen 1
|
|
chr9_+_56983627
Show fit
|
0.16 |
ENSMUST00000168678.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast)
|
|
chr17_-_51486196
Show fit
|
0.16 |
ENSMUST00000024717.10
ENSMUST00000224528.2
|
Tbc1d5
|
TBC1 domain family, member 5
|
|
chr19_+_6468761
Show fit
|
0.16 |
ENSMUST00000113462.8
ENSMUST00000077182.13
ENSMUST00000236635.2
ENSMUST00000113461.8
|
Nrxn2
|
neurexin II
|
|
chr16_-_90081300
Show fit
|
0.16 |
ENSMUST00000039280.9
ENSMUST00000232371.2
|
Scaf4
|
SR-related CTD-associated factor 4
|
|
chrX_+_137815171
Show fit
|
0.16 |
ENSMUST00000064937.14
ENSMUST00000113052.8
|
Nrk
|
Nik related kinase
|
|
chr5_+_147367237
Show fit
|
0.16 |
ENSMUST00000176600.8
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit
|
|
chr1_+_156386327
Show fit
|
0.16 |
ENSMUST00000173929.8
|
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene)
|
|
chr15_-_76084035
Show fit
|
0.16 |
ENSMUST00000054449.14
ENSMUST00000169714.8
ENSMUST00000165453.8
|
Plec
|
plectin
|
|
chr8_+_27467330
Show fit
|
0.16 |
ENSMUST00000127097.9
ENSMUST00000154256.3
|
Zfp703
|
zinc finger protein 703
|
|
chr11_+_74721733
Show fit
|
0.16 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein
|
|
chr1_-_123973223
Show fit
|
0.16 |
ENSMUST00000112606.8
|
Dpp10
|
dipeptidylpeptidase 10
|
|
chr7_-_78228116
Show fit
|
0.16 |
ENSMUST00000206268.2
ENSMUST00000039431.14
|
Ntrk3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chr2_-_160714904
Show fit
|
0.15 |
ENSMUST00000109460.8
ENSMUST00000127201.2
|
Zhx3
|
zinc fingers and homeoboxes 3
|
|
chr7_+_27879650
Show fit
|
0.15 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b
|
|
chr13_-_111945499
Show fit
|
0.15 |
ENSMUST00000109267.9
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr19_+_10502612
Show fit
|
0.15 |
ENSMUST00000237321.2
ENSMUST00000038379.5
|
Cpsf7
|
cleavage and polyadenylation specific factor 7
|
|
chr7_-_98305737
Show fit
|
0.15 |
ENSMUST00000205911.2
ENSMUST00000038359.6
ENSMUST00000206611.2
ENSMUST00000206619.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor
|
|
chr19_-_5713701
Show fit
|
0.15 |
ENSMUST00000164304.9
ENSMUST00000237544.2
|
Sipa1
|
signal-induced proliferation associated gene 1
|
|
chr7_-_25454177
Show fit
|
0.15 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr2_-_38177359
Show fit
|
0.15 |
ENSMUST00000102787.10
|
Dennd1a
|
DENN/MADD domain containing 1A
|
|
chr2_-_60711706
Show fit
|
0.15 |
ENSMUST00000164147.8
ENSMUST00000112509.2
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr1_-_52766615
Show fit
|
0.15 |
ENSMUST00000156876.8
ENSMUST00000087701.4
|
Mfsd6
|
major facilitator superfamily domain containing 6
|
|
chr9_+_56983679
Show fit
|
0.15 |
ENSMUST00000168177.8
|
Sin3a
|
transcriptional regulator, SIN3A (yeast)
|
|
chr11_-_86248395
Show fit
|
0.15 |
ENSMUST00000043624.9
|
Med13
|
mediator complex subunit 13
|
|
chr16_-_10131804
Show fit
|
0.15 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2
|
|
chr13_-_54835996
Show fit
|
0.15 |
ENSMUST00000150806.8
ENSMUST00000125927.8
|
Rnf44
|
ring finger protein 44
|
|
chr9_+_87026337
Show fit
|
0.14 |
ENSMUST00000113149.8
ENSMUST00000049457.14
ENSMUST00000179313.3
|
Mrap2
|
melanocortin 2 receptor accessory protein 2
|
|
chr14_+_99536111
Show fit
|
0.14 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5
|
|
chr19_-_10847121
Show fit
|
0.14 |
ENSMUST00000120524.2
ENSMUST00000025645.14
|
Tmem132a
|
transmembrane protein 132A
|
|
chr13_-_58261406
Show fit
|
0.14 |
ENSMUST00000160860.9
|
Klhl3
|
kelch-like 3
|
|
chr10_-_66932615
Show fit
|
0.14 |
ENSMUST00000217841.2
|
Reep3
|
receptor accessory protein 3
|
|
chr2_-_172782089
Show fit
|
0.14 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7
|
|
chr14_+_20979466
Show fit
|
0.14 |
ENSMUST00000022369.9
|
Vcl
|
vinculin
|
|
chrX_-_72868544
Show fit
|
0.14 |
ENSMUST00000002080.12
ENSMUST00000114438.3
|
Pdzd4
|
PDZ domain containing 4
|
|
chr4_-_126096112
Show fit
|
0.14 |
ENSMUST00000142125.2
ENSMUST00000106141.3
|
Thrap3
|
thyroid hormone receptor associated protein 3
|
|
chr2_+_157870399
Show fit
|
0.14 |
ENSMUST00000103123.10
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr5_-_108017019
Show fit
|
0.14 |
ENSMUST00000128723.8
ENSMUST00000124034.8
|
Evi5
|
ecotropic viral integration site 5
|
|
chr7_+_100355910
Show fit
|
0.13 |
ENSMUST00000207875.2
ENSMUST00000208013.2
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr18_+_7869370
Show fit
|
0.13 |
ENSMUST00000092112.11
ENSMUST00000172018.8
ENSMUST00000168446.8
|
Wac
|
WW domain containing adaptor with coiled-coil
|
|
chr3_-_129834788
Show fit
|
0.13 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae)
|
|
chr7_-_98305986
Show fit
|
0.13 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor
|
|
chr5_+_53747796
Show fit
|
0.13 |
ENSMUST00000113865.5
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr7_+_26895206
Show fit
|
0.13 |
ENSMUST00000179391.8
ENSMUST00000108379.8
|
BC024978
|
cDNA sequence BC024978
|
|
chr10_+_66932235
Show fit
|
0.13 |
ENSMUST00000174317.8
|
Jmjd1c
|
jumonji domain containing 1C
|
|
chr17_-_57137898
Show fit
|
0.13 |
ENSMUST00000233000.2
ENSMUST00000002444.15
ENSMUST00000086801.7
|
Rfx2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr10_+_110581293
Show fit
|
0.13 |
ENSMUST00000174857.8
ENSMUST00000073781.12
ENSMUST00000173471.8
ENSMUST00000173634.2
|
E2f7
|
E2F transcription factor 7
|
|
chr4_+_122889737
Show fit
|
0.13 |
ENSMUST00000106252.9
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived
|
|
chr9_-_58648826
Show fit
|
0.13 |
ENSMUST00000098674.6
|
Rec114
|
REC114 meiotic recombination protein
|
|
chr15_-_89239858
Show fit
|
0.13 |
ENSMUST00000023283.6
|
Lmf2
|
lipase maturation factor 2
|
|
chr2_-_126985226
Show fit
|
0.13 |
ENSMUST00000110386.2
ENSMUST00000132773.2
|
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1
|
|
chr6_+_42326528
Show fit
|
0.13 |
ENSMUST00000203329.3
|
Zyx
|
zyxin
|
|
chr13_-_54835878
Show fit
|
0.13 |
ENSMUST00000125871.8
|
Rnf44
|
ring finger protein 44
|
|
chr5_-_106844685
Show fit
|
0.13 |
ENSMUST00000127434.8
ENSMUST00000112696.8
ENSMUST00000112698.8
|
Zfp644
|
zinc finger protein 644
|
|
chr16_+_27207722
Show fit
|
0.13 |
ENSMUST00000039443.14
ENSMUST00000096127.11
|
Ccdc50
|
coiled-coil domain containing 50
|
|
chr17_-_56891576
Show fit
|
0.12 |
ENSMUST00000075510.12
|
Safb2
|
scaffold attachment factor B2
|
|
chr16_+_33504908
Show fit
|
0.12 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1
|
|
chr15_-_77726333
Show fit
|
0.12 |
ENSMUST00000016771.13
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle
|
|
chr1_-_64776890
Show fit
|
0.12 |
ENSMUST00000116133.4
ENSMUST00000063982.7
|
Fzd5
|
frizzled class receptor 5
|
|
chr15_-_97991114
Show fit
|
0.12 |
ENSMUST00000180657.2
|
Senp1
|
SUMO1/sentrin specific peptidase 1
|
|
chr3_-_88366159
Show fit
|
0.12 |
ENSMUST00000147200.8
ENSMUST00000169222.8
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr10_-_7162196
Show fit
|
0.12 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3
|
|
chr12_+_16945118
Show fit
|
0.12 |
ENSMUST00000220688.2
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2
|
|
chr6_-_48422307
Show fit
|
0.12 |
ENSMUST00000114563.8
ENSMUST00000114558.8
ENSMUST00000101443.10
|
Zfp467
|
zinc finger protein 467
|
|
chr11_+_70431063
Show fit
|
0.12 |
ENSMUST00000018429.12
ENSMUST00000108557.10
ENSMUST00000108556.2
|
Pld2
|
phospholipase D2
|
|
chr10_+_105676925
Show fit
|
0.12 |
ENSMUST00000020049.9
|
Ccdc59
|
coiled-coil domain containing 59
|
|
chr11_-_5049036
Show fit
|
0.12 |
ENSMUST00000102930.10
ENSMUST00000093365.12
ENSMUST00000073308.11
|
Ewsr1
|
Ewing sarcoma breakpoint region 1
|
|
chr3_+_137573436
Show fit
|
0.12 |
ENSMUST00000090178.10
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14
|
|
chr7_+_119495058
Show fit
|
0.12 |
ENSMUST00000106518.9
ENSMUST00000207270.2
ENSMUST00000208424.2
ENSMUST00000208202.2
ENSMUST00000054440.11
|
Lyrm1
|
LYR motif containing 1
|
|
chr10_+_107998033
Show fit
|
0.12 |
ENSMUST00000219263.2
|
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr18_+_36693646
Show fit
|
0.12 |
ENSMUST00000155329.9
|
Ankhd1
|
ankyrin repeat and KH domain containing 1
|
|
chr3_+_137573839
Show fit
|
0.12 |
ENSMUST00000197711.2
|
Dnajb14
|
DnaJ heat shock protein family (Hsp40) member B14
|
|
chr12_-_100691316
Show fit
|
0.12 |
ENSMUST00000222731.2
|
Rps6ka5
|
ribosomal protein S6 kinase, polypeptide 5
|
|
chr6_-_122317156
Show fit
|
0.12 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1
|
|
chr4_+_122889828
Show fit
|
0.12 |
ENSMUST00000030407.8
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived
|
|
chr15_-_102165884
Show fit
|
0.12 |
ENSMUST00000043172.15
|
Rarg
|
retinoic acid receptor, gamma
|
|
chr1_-_13442658
Show fit
|
0.12 |
ENSMUST00000081713.11
|
Ncoa2
|
nuclear receptor coactivator 2
|
|
chrX_+_72454702
Show fit
|
0.12 |
ENSMUST00000033740.12
|
Zfp92
|
zinc finger protein 92
|
|
chr2_+_146063841
Show fit
|
0.11 |
ENSMUST00000089257.6
|
Insm1
|
insulinoma-associated 1
|
|
chr7_-_29426425
Show fit
|
0.11 |
ENSMUST00000061193.4
|
Catsperg2
|
cation channel sperm associated auxiliary subunit gamma 2
|
|
chr16_-_90808196
Show fit
|
0.11 |
ENSMUST00000121759.8
|
Synj1
|
synaptojanin 1
|
|
chr16_+_57369595
Show fit
|
0.11 |
ENSMUST00000159414.2
|
Filip1l
|
filamin A interacting protein 1-like
|
|
chr10_+_111342147
Show fit
|
0.11 |
ENSMUST00000164773.2
|
Phlda1
|
pleckstrin homology like domain, family A, member 1
|
|
chr9_-_21159733
Show fit
|
0.11 |
ENSMUST00000122088.2
|
S1pr5
|
sphingosine-1-phosphate receptor 5
|
|
chr2_-_155676765
Show fit
|
0.11 |
ENSMUST00000029143.7
ENSMUST00000239423.2
|
Fam83c
|
family with sequence similarity 83, member C
|
|
chr11_-_76969230
Show fit
|
0.11 |
ENSMUST00000102494.8
|
Nsrp1
|
nuclear speckle regulatory protein 1
|
|
chr7_+_128290204
Show fit
|
0.11 |
ENSMUST00000118605.2
|
Inpp5f
|
inositol polyphosphate-5-phosphatase F
|
|
chr15_-_102154874
Show fit
|
0.11 |
ENSMUST00000063339.14
|
Rarg
|
retinoic acid receptor, gamma
|
|
chr7_-_25454126
Show fit
|
0.11 |
ENSMUST00000108401.3
ENSMUST00000043765.14
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1
|
|
chr2_-_31031814
Show fit
|
0.11 |
ENSMUST00000073879.12
ENSMUST00000100208.9
ENSMUST00000100207.9
ENSMUST00000113555.8
ENSMUST00000075326.11
ENSMUST00000113552.9
ENSMUST00000136181.8
ENSMUST00000113564.9
ENSMUST00000113562.9
ENSMUST00000113560.8
|
Fnbp1
|
formin binding protein 1
|
|
chr2_+_121697398
Show fit
|
0.11 |
ENSMUST00000110586.10
ENSMUST00000078752.10
|
Golm2
|
golgi membrane protein 2
|
|
chr15_-_78004211
Show fit
|
0.11 |
ENSMUST00000019290.3
|
Cacng2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr2_-_130748259
Show fit
|
0.11 |
ENSMUST00000128176.2
ENSMUST00000133766.2
ENSMUST00000135110.2
ENSMUST00000146975.2
|
A730017L22Rik
4930402H24Rik
|
RIKEN cDNA A730017L22 gene
RIKEN cDNA 4930402H24 gene
|
|
chrX_-_73009933
Show fit
|
0.11 |
ENSMUST00000114372.3
ENSMUST00000033761.13
|
Hcfc1
|
host cell factor C1
|
|
chr15_+_12117899
Show fit
|
0.11 |
ENSMUST00000122941.8
|
Zfr
|
zinc finger RNA binding protein
|
|
chr5_-_90514061
Show fit
|
0.11 |
ENSMUST00000081914.13
|
Ankrd17
|
ankyrin repeat domain 17
|
|
chrX_-_72830487
Show fit
|
0.11 |
ENSMUST00000052761.9
|
Idh3g
|
isocitrate dehydrogenase 3 (NAD+), gamma
|
|
chrX_-_132882514
Show fit
|
0.11 |
ENSMUST00000113297.9
ENSMUST00000174542.2
ENSMUST00000033608.15
ENSMUST00000113294.8
|
Sytl4
|
synaptotagmin-like 4
|
|
chr2_-_38177182
Show fit
|
0.11 |
ENSMUST00000130472.8
|
Dennd1a
|
DENN/MADD domain containing 1A
|
|
chr18_+_34354031
Show fit
|
0.11 |
ENSMUST00000115781.10
ENSMUST00000079362.13
|
Apc
|
APC, WNT signaling pathway regulator
|
|
chr5_+_123280250
Show fit
|
0.11 |
ENSMUST00000174836.8
ENSMUST00000163030.9
|
Setd1b
|
SET domain containing 1B
|
|
chr17_+_34251041
Show fit
|
0.11 |
ENSMUST00000173354.9
|
Rxrb
|
retinoid X receptor beta
|
|
chr11_-_97590460
Show fit
|
0.11 |
ENSMUST00000103148.8
ENSMUST00000169807.8
|
Pcgf2
|
polycomb group ring finger 2
|
|
chr11_-_69649004
Show fit
|
0.11 |
ENSMUST00000071213.4
|
Polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A
|
|
chr10_+_70010839
Show fit
|
0.11 |
ENSMUST00000156001.8
ENSMUST00000135607.2
|
Ccdc6
|
coiled-coil domain containing 6
|
|
chr4_+_129229373
Show fit
|
0.11 |
ENSMUST00000141235.8
|
Zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand
|
|
chr4_+_84802592
Show fit
|
0.11 |
ENSMUST00000102819.10
|
Cntln
|
centlein, centrosomal protein
|
|
chr2_+_54326329
Show fit
|
0.11 |
ENSMUST00000112636.8
ENSMUST00000112635.8
ENSMUST00000112634.8
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13
|
|
chr10_-_127456791
Show fit
|
0.11 |
ENSMUST00000118455.2
ENSMUST00000121829.8
|
Lrp1
|
low density lipoprotein receptor-related protein 1
|
|
chr8_+_27575611
Show fit
|
0.11 |
ENSMUST00000178514.8
ENSMUST00000033876.14
|
Adgra2
|
adhesion G protein-coupled receptor A2
|
|
chr15_-_81074921
Show fit
|
0.11 |
ENSMUST00000131235.9
ENSMUST00000134469.9
ENSMUST00000239114.2
ENSMUST00000149582.8
|
Mrtfa
|
myocardin related transcription factor A
|
|
chr5_+_124577952
Show fit
|
0.11 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A
|
|
chr19_-_5323092
Show fit
|
0.11 |
ENSMUST00000237463.2
ENSMUST00000025786.9
|
Pacs1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr5_-_90514360
Show fit
|
0.11 |
ENSMUST00000168058.7
ENSMUST00000014421.15
|
Ankrd17
|
ankyrin repeat domain 17
|
|
chr13_-_105191403
Show fit
|
0.11 |
ENSMUST00000063551.7
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein
|
|
chr9_+_61279640
Show fit
|
0.11 |
ENSMUST00000178113.8
ENSMUST00000159386.8
|
Tle3
|
transducin-like enhancer of split 3
|
|
chr15_+_82225380
Show fit
|
0.11 |
ENSMUST00000050349.3
|
Pheta2
|
PH domain containing endocytic trafficking adaptor 2
|
|
chr11_-_102446947
Show fit
|
0.11 |
ENSMUST00000143842.2
|
Gpatch8
|
G patch domain containing 8
|
|
chr11_+_49094119
Show fit
|
0.11 |
ENSMUST00000109198.8
ENSMUST00000137061.9
|
Zfp62
|
zinc finger protein 62
|
|
chr16_-_18066591
Show fit
|
0.11 |
ENSMUST00000115645.10
|
Ranbp1
|
RAN binding protein 1
|
|
chr7_+_127111576
Show fit
|
0.10 |
ENSMUST00000186672.7
|
Srcap
|
Snf2-related CREBBP activator protein
|