Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
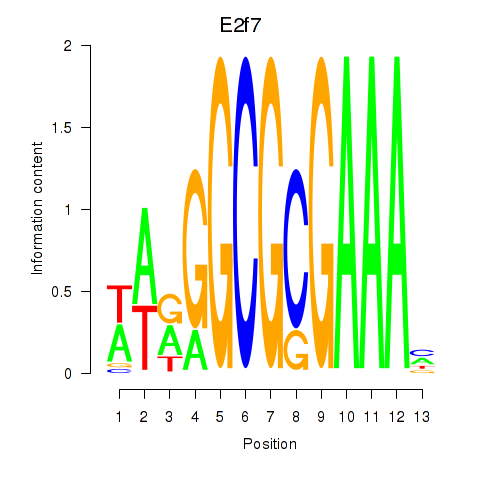
Results for E2f7
Z-value: 3.16
Transcription factors associated with E2f7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f7
|
ENSMUSG00000020185.17 | E2F transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f7 | mm39_v1_chr10_+_110581293_110581433 | 0.52 | 3.7e-01 | Click! |
Activity profile of E2f7 motif
Sorted Z-values of E2f7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_21938258 | 9.08 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chr13_-_21937997 | 7.99 |
ENSMUST00000074752.4
|
H2ac15
|
H2A clustered histone 15 |
| chr13_+_22017906 | 7.02 |
ENSMUST00000180288.2
|
H2bc24
|
H2B clustered histone 24 |
| chr13_+_22220000 | 6.96 |
ENSMUST00000110455.4
|
H2bc12
|
H2B clustered histone 12 |
| chr13_-_23758370 | 6.92 |
ENSMUST00000105106.2
|
H2bc7
|
H2B clustered histone 7 |
| chr13_+_22227359 | 6.67 |
ENSMUST00000110452.2
|
H2bc11
|
H2B clustered histone 11 |
| chr13_-_21967540 | 6.12 |
ENSMUST00000189457.2
|
H3c11
|
H3 clustered histone 11 |
| chr13_-_22219738 | 4.75 |
ENSMUST00000091742.6
|
H2ac12
|
H2A clustered histone 12 |
| chr13_-_21900313 | 4.72 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13 |
| chr13_-_21994366 | 4.70 |
ENSMUST00000091749.4
|
H2bc23
|
H2B clustered histone 23 |
| chr13_-_21934675 | 4.50 |
ENSMUST00000102983.2
|
H4c12
|
H4 clustered histone 12 |
| chr9_+_3013140 | 4.10 |
ENSMUST00000143083.3
|
Gm10721
|
predicted gene 10721 |
| chr9_+_3025417 | 3.45 |
ENSMUST00000075573.7
|
Gm10717
|
predicted gene 10717 |
| chr13_-_22017677 | 3.37 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr13_-_22227114 | 3.26 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chr9_+_3017408 | 3.13 |
ENSMUST00000099049.4
|
Gm10719
|
predicted gene 10719 |
| chr13_+_21994588 | 2.94 |
ENSMUST00000091745.6
|
H2ac23
|
H2A clustered histone 23 |
| chr13_+_21900554 | 2.87 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13 |
| chr9_+_3037111 | 2.12 |
ENSMUST00000177969.2
|
Gm10715
|
predicted gene 10715 |
| chr13_+_23758555 | 2.11 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chr9_+_3015654 | 2.04 |
ENSMUST00000099050.4
|
Gm10720
|
predicted gene 10720 |
| chrX_-_73009933 | 1.32 |
ENSMUST00000114372.3
ENSMUST00000033761.13 |
Hcfc1
|
host cell factor C1 |
| chr7_-_25454177 | 1.25 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr15_-_58953838 | 1.25 |
ENSMUST00000080371.8
|
Mtss1
|
MTSS I-BAR domain containing 1 |
| chr7_-_98305737 | 1.23 |
ENSMUST00000205911.2
ENSMUST00000038359.6 ENSMUST00000206611.2 ENSMUST00000206619.2 |
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr12_-_44257109 | 1.19 |
ENSMUST00000015049.5
|
Dnajb9
|
DnaJ heat shock protein family (Hsp40) member B9 |
| chr7_+_3648264 | 1.04 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr2_+_27776428 | 0.96 |
ENSMUST00000028280.14
|
Col5a1
|
collagen, type V, alpha 1 |
| chr7_+_44465714 | 0.96 |
ENSMUST00000208172.2
|
Nup62
|
nucleoporin 62 |
| chr7_-_25454126 | 0.94 |
ENSMUST00000108401.3
ENSMUST00000043765.14 |
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr9_-_121106209 | 0.90 |
ENSMUST00000051479.13
ENSMUST00000171923.8 |
Ulk4
|
unc-51-like kinase 4 |
| chr14_-_54491365 | 0.90 |
ENSMUST00000128231.2
|
Dad1
|
defender against cell death 1 |
| chr7_-_98305986 | 0.88 |
ENSMUST00000205276.2
|
Emsy
|
EMSY, BRCA2-interacting transcriptional repressor |
| chr1_-_74544946 | 0.81 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr12_+_11316101 | 0.73 |
ENSMUST00000218866.2
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr7_-_28297565 | 0.72 |
ENSMUST00000040531.9
ENSMUST00000108283.8 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 (RAC1) activated kinase 4 |
| chr11_+_69214789 | 0.69 |
ENSMUST00000102602.8
|
Trappc1
|
trafficking protein particle complex 1 |
| chr11_+_98441923 | 0.66 |
ENSMUST00000081033.13
ENSMUST00000107511.8 ENSMUST00000107509.8 ENSMUST00000017339.12 |
Zpbp2
|
zona pellucida binding protein 2 |
| chr11_+_69214971 | 0.61 |
ENSMUST00000108662.2
|
Trappc1
|
trafficking protein particle complex 1 |
| chr10_-_125225298 | 0.60 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7 |
| chrX_+_93278588 | 0.60 |
ENSMUST00000096369.10
ENSMUST00000113911.9 |
Klhl15
|
kelch-like 15 |
| chr15_+_82183143 | 0.55 |
ENSMUST00000023089.5
|
Wbp2nl
|
WBP2 N-terminal like |
| chr6_-_30509737 | 0.51 |
ENSMUST00000154547.3
ENSMUST00000115160.10 |
Tmem209
|
transmembrane protein 209 |
| chr11_+_69214883 | 0.51 |
ENSMUST00000102601.10
|
Trappc1
|
trafficking protein particle complex 1 |
| chr13_-_97334859 | 0.49 |
ENSMUST00000022169.10
|
Hexb
|
hexosaminidase B |
| chr9_-_87137515 | 0.46 |
ENSMUST00000093802.6
|
Cep162
|
centrosomal protein 162 |
| chrX_+_67722230 | 0.43 |
ENSMUST00000114656.8
|
Fmr1
|
FMRP translational regulator 1 |
| chr1_+_88034556 | 0.43 |
ENSMUST00000113137.2
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B |
| chr15_-_97991114 | 0.42 |
ENSMUST00000180657.2
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr11_+_69214895 | 0.42 |
ENSMUST00000060956.13
|
Trappc1
|
trafficking protein particle complex 1 |
| chrX_+_93278526 | 0.41 |
ENSMUST00000113908.8
ENSMUST00000113916.10 |
Klhl15
|
kelch-like 15 |
| chr1_+_172348611 | 0.38 |
ENSMUST00000085894.12
ENSMUST00000161140.8 ENSMUST00000162988.8 |
Cfap45
|
cilia and flagella associated protein 45 |
| chr12_+_73333553 | 0.37 |
ENSMUST00000140523.8
ENSMUST00000126488.8 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr17_-_35265514 | 0.36 |
ENSMUST00000007250.14
|
Msh5
|
mutS homolog 5 |
| chr14_-_66071337 | 0.34 |
ENSMUST00000225853.2
|
Esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr3_+_88049633 | 0.31 |
ENSMUST00000001455.13
ENSMUST00000119251.8 |
Mef2d
|
myocyte enhancer factor 2D |
| chr3_+_60408678 | 0.31 |
ENSMUST00000191747.6
ENSMUST00000194069.6 |
Mbnl1
|
muscleblind like splicing factor 1 |
| chr4_+_126450762 | 0.31 |
ENSMUST00000147675.2
|
Clspn
|
claspin |
| chr3_+_103187162 | 0.29 |
ENSMUST00000106860.6
|
Trim33
|
tripartite motif-containing 33 |
| chr1_-_88629843 | 0.27 |
ENSMUST00000159814.2
|
Arl4c
|
ADP-ribosylation factor-like 4C |
| chr6_+_113054592 | 0.27 |
ENSMUST00000113157.8
|
Setd5
|
SET domain containing 5 |
| chr17_-_35340966 | 0.25 |
ENSMUST00000173915.3
ENSMUST00000172765.9 |
Csnk2b
|
casein kinase 2, beta polypeptide |
| chr6_-_52160816 | 0.24 |
ENSMUST00000134831.2
|
Hoxa3
|
homeobox A3 |
| chr7_+_44465806 | 0.23 |
ENSMUST00000207103.2
ENSMUST00000118125.9 |
Nup62
Il4i1
|
nucleoporin 62 interleukin 4 induced 1 |
| chr13_-_47259652 | 0.19 |
ENSMUST00000021807.13
ENSMUST00000135278.8 |
Dek
|
DEK proto-oncogene (DNA binding) |
| chr6_-_30509705 | 0.18 |
ENSMUST00000064330.13
ENSMUST00000102991.9 ENSMUST00000115157.8 ENSMUST00000148638.2 |
Tmem209
|
transmembrane protein 209 |
| chr3_+_88439616 | 0.15 |
ENSMUST00000172699.2
|
Mex3a
|
mex3 RNA binding family member A |
| chr8_+_26008773 | 0.14 |
ENSMUST00000084027.13
ENSMUST00000178276.8 ENSMUST00000179592.8 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chr2_-_35994072 | 0.12 |
ENSMUST00000112961.10
ENSMUST00000112966.10 |
Lhx6
|
LIM homeobox protein 6 |
| chr17_-_35069136 | 0.12 |
ENSMUST00000046022.16
|
Skiv2l
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr10_+_67374366 | 0.12 |
ENSMUST00000127820.2
|
Egr2
|
early growth response 2 |
| chr1_+_153300874 | 0.09 |
ENSMUST00000042373.12
|
Shcbp1l
|
Shc SH2-domain binding protein 1-like |
| chr1_-_190915441 | 0.09 |
ENSMUST00000027941.14
|
Atf3
|
activating transcription factor 3 |
| chr6_+_30509826 | 0.08 |
ENSMUST00000031797.11
|
Ssmem1
|
serine-rich single-pass membrane protein 1 |
| chr19_-_41195213 | 0.08 |
ENSMUST00000169941.2
ENSMUST00000025986.15 |
Tll2
|
tolloid-like 2 |
| chr14_-_17398733 | 0.07 |
ENSMUST00000163719.8
|
Gm8281
|
predicted gene, 8281 |
| chrX_+_6690410 | 0.07 |
ENSMUST00000145302.3
|
Dgkk
|
diacylglycerol kinase kappa |
| chr16_+_76810588 | 0.07 |
ENSMUST00000239066.2
ENSMUST00000023580.8 |
Usp25
|
ubiquitin specific peptidase 25 |
| chr2_-_65194344 | 0.06 |
ENSMUST00000155962.3
ENSMUST00000112420.8 ENSMUST00000152324.8 |
Slc38a11
|
solute carrier family 38, member 11 |
| chr4_+_129000600 | 0.05 |
ENSMUST00000148979.2
|
Tmem54
|
transmembrane protein 54 |
| chr11_+_85202058 | 0.04 |
ENSMUST00000020835.16
|
Ppm1d
|
protein phosphatase 1D magnesium-dependent, delta isoform |
| chr12_+_11506053 | 0.04 |
ENSMUST00000124065.2
|
Rad51ap2
|
RAD51 associated protein 2 |
| chr4_+_94627755 | 0.04 |
ENSMUST00000071168.6
|
Tek
|
TEK receptor tyrosine kinase |
| chr9_+_46194293 | 0.04 |
ENSMUST00000074957.5
|
Bud13
|
BUD13 homolog |
| chr14_-_18287197 | 0.04 |
ENSMUST00000164512.8
|
Gm2974
|
predicted gene 2974 |
| chr4_+_33062999 | 0.04 |
ENSMUST00000108162.8
ENSMUST00000024035.9 |
Gabrr2
|
gamma-aminobutyric acid (GABA) C receptor, subunit rho 2 |
| chr2_+_23211258 | 0.04 |
ENSMUST00000102945.2
|
Nxph2
|
neurexophilin 2 |
| chr4_+_119280002 | 0.04 |
ENSMUST00000094819.5
|
Zmynd12
|
zinc finger, MYND domain containing 12 |
| chr14_-_17742998 | 0.03 |
ENSMUST00000165619.8
|
Gm3252
|
predicted gene 3252 |
| chr14_-_18897750 | 0.03 |
ENSMUST00000178728.2
|
Gm3005
|
predicted gene 3005 |
| chr4_-_44703413 | 0.03 |
ENSMUST00000186542.2
|
Pax5
|
paired box 5 |
| chr1_-_135095344 | 0.02 |
ENSMUST00000027682.9
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr14_-_18359247 | 0.02 |
ENSMUST00000170207.8
|
Gm8108
|
predicted gene 8108 |
| chr14_+_15579811 | 0.02 |
ENSMUST00000171906.2
|
Gm3667
|
predicted gene 3667 |
| chr14_+_15295240 | 0.02 |
ENSMUST00000172431.8
|
Gm3512
|
predicted gene 3512 |
| chr14_-_17614197 | 0.02 |
ENSMUST00000166776.8
|
Gm3264
|
predicted gene 3264 |
| chr14_+_15442324 | 0.02 |
ENSMUST00000170738.3
|
Gm10406
|
predicted gene 10406 |
| chr2_+_172392678 | 0.02 |
ENSMUST00000099058.10
|
Tfap2c
|
transcription factor AP-2, gamma |
| chr19_-_11806388 | 0.02 |
ENSMUST00000061235.3
|
Olfr1417
|
olfactory receptor 1417 |
| chr19_+_16110176 | 0.02 |
ENSMUST00000025541.6
|
Gnaq
|
guanine nucleotide binding protein, alpha q polypeptide |
| chr8_+_26008799 | 0.02 |
ENSMUST00000119398.10
ENSMUST00000117179.9 |
Fgfr1
|
fibroblast growth factor receptor 1 |
| chrX_+_73637934 | 0.02 |
ENSMUST00000054541.3
|
Olfr1325
|
olfactory receptor 1325 |
| chr14_-_18659699 | 0.01 |
ENSMUST00000170480.8
|
Gm3002
|
predicted gene 3002 |
| chr13_-_55477535 | 0.01 |
ENSMUST00000021941.8
|
Mxd3
|
Max dimerization protein 3 |
| chr11_-_106192627 | 0.01 |
ENSMUST00000103071.4
|
Gh
|
growth hormone |
| chr11_+_49039086 | 0.01 |
ENSMUST00000059379.2
|
Olfr1395
|
olfactory receptor 1395 |
| chr12_+_11315868 | 0.01 |
ENSMUST00000020931.6
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr16_-_37205302 | 0.00 |
ENSMUST00000114781.8
ENSMUST00000114780.8 |
Stxbp5l
|
syntaxin binding protein 5-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.2 | 1.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.7 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.2 | 1.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 1.2 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.1 | 0.4 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.1 | 1.0 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.6 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:0006689 | ganglioside catabolic process(GO:0006689) penetration of zona pellucida(GO:0007341) oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.2 | GO:1903465 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.0 | 0.9 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:0021615 | specification of organ position(GO:0010159) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 0.4 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.7 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 1.1 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.7 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.4 | GO:1902737 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.1 | 2.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.3 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.9 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.6 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 1.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |