Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
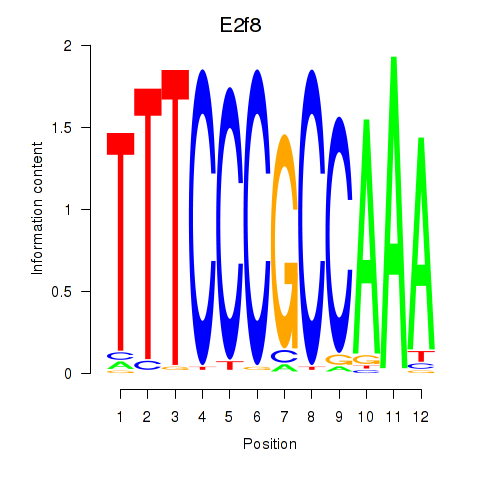
Results for E2f8
Z-value: 2.61
Transcription factors associated with E2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f8
|
ENSMUSG00000046179.18 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f8 | mm39_v1_chr7_-_48531344_48531367 | -0.87 | 5.6e-02 | Click! |
Activity profile of E2f8 motif
Sorted Z-values of E2f8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_75836187 | 2.82 |
ENSMUST00000164309.3
ENSMUST00000212426.2 ENSMUST00000212811.2 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr12_+_24758968 | 2.37 |
ENSMUST00000154588.2
|
Rrm2
|
ribonucleotide reductase M2 |
| chr12_+_24758724 | 2.37 |
ENSMUST00000153058.8
|
Rrm2
|
ribonucleotide reductase M2 |
| chr1_-_128287347 | 2.35 |
ENSMUST00000190495.2
ENSMUST00000027601.11 |
Mcm6
|
minichromosome maintenance complex component 6 |
| chr2_+_162896602 | 2.26 |
ENSMUST00000018005.10
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr3_-_90603013 | 1.62 |
ENSMUST00000069960.12
ENSMUST00000117167.2 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr15_+_97990431 | 1.61 |
ENSMUST00000229280.2
ENSMUST00000163507.8 ENSMUST00000230445.2 |
Pfkm
|
phosphofructokinase, muscle |
| chr12_-_55045887 | 1.54 |
ENSMUST00000173529.2
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr11_+_87938626 | 1.49 |
ENSMUST00000107920.10
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr4_+_132495636 | 1.42 |
ENSMUST00000102561.11
|
Rpa2
|
replication protein A2 |
| chr11_-_101676076 | 1.39 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr7_+_101714943 | 1.29 |
ENSMUST00000094130.4
ENSMUST00000084843.10 |
Xndc1
Xntrpc
|
Xrcc1 N-terminal domain containing 1 Xndc1-transient receptor potential cation channel, subfamily C, member 2 readthrough |
| chr17_+_56610321 | 1.25 |
ENSMUST00000001258.15
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr9_-_36637670 | 1.25 |
ENSMUST00000172702.9
ENSMUST00000172742.2 |
Chek1
|
checkpoint kinase 1 |
| chr7_-_44198157 | 1.19 |
ENSMUST00000145956.2
ENSMUST00000049343.15 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr11_+_79883885 | 1.16 |
ENSMUST00000163272.2
ENSMUST00000017692.15 |
Suz12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr6_+_125016723 | 1.16 |
ENSMUST00000140131.8
ENSMUST00000032480.14 |
Ing4
|
inhibitor of growth family, member 4 |
| chr12_+_55883101 | 1.15 |
ENSMUST00000059250.8
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr11_+_87938128 | 1.11 |
ENSMUST00000139129.9
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr9_-_36637923 | 1.08 |
ENSMUST00000034625.12
|
Chek1
|
checkpoint kinase 1 |
| chr17_+_56610396 | 1.07 |
ENSMUST00000113038.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr9_-_50571080 | 1.04 |
ENSMUST00000034567.4
|
Dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr2_-_34803988 | 1.03 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr11_+_87938519 | 1.03 |
ENSMUST00000079866.11
|
Srsf1
|
serine and arginine-rich splicing factor 1 |
| chr6_-_88875646 | 1.01 |
ENSMUST00000058011.8
|
Mcm2
|
minichromosome maintenance complex component 2 |
| chr15_-_55421144 | 0.93 |
ENSMUST00000172387.8
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr7_+_101714692 | 0.89 |
ENSMUST00000106950.8
ENSMUST00000146450.8 |
Xndc1
|
Xrcc1 N-terminal domain containing 1 |
| chr17_+_56347424 | 0.88 |
ENSMUST00000002914.10
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr6_+_35154319 | 0.83 |
ENSMUST00000201374.4
ENSMUST00000043815.16 |
Nup205
|
nucleoporin 205 |
| chr5_+_123887759 | 0.83 |
ENSMUST00000031366.12
|
Kntc1
|
kinetochore associated 1 |
| chr10_+_127851031 | 0.82 |
ENSMUST00000178041.8
ENSMUST00000026461.8 |
Prim1
|
DNA primase, p49 subunit |
| chr17_+_34129221 | 0.81 |
ENSMUST00000174541.9
|
Daxx
|
Fas death domain-associated protein |
| chr17_+_88282472 | 0.77 |
ENSMUST00000005503.5
|
Msh6
|
mutS homolog 6 |
| chr15_+_101071948 | 0.77 |
ENSMUST00000000544.12
|
Acvr1b
|
activin A receptor, type 1B |
| chr2_-_157046386 | 0.71 |
ENSMUST00000029170.8
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr5_-_137785903 | 0.68 |
ENSMUST00000196022.2
|
Mepce
|
methylphosphate capping enzyme |
| chr19_+_38919353 | 0.67 |
ENSMUST00000025965.12
|
Hells
|
helicase, lymphoid specific |
| chr4_+_135583018 | 0.62 |
ENSMUST00000105853.10
ENSMUST00000097844.9 ENSMUST00000102544.9 ENSMUST00000126641.2 |
Srsf10
|
serine and arginine-rich splicing factor 10 |
| chr11_-_21521934 | 0.61 |
ENSMUST00000239073.2
|
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr11_-_21522193 | 0.59 |
ENSMUST00000102874.11
ENSMUST00000238916.2 |
Mdh1
|
malate dehydrogenase 1, NAD (soluble) |
| chr7_+_109721230 | 0.58 |
ENSMUST00000033326.10
|
Wee1
|
WEE 1 homolog 1 (S. pombe) |
| chr6_+_117894242 | 0.58 |
ENSMUST00000180020.8
ENSMUST00000177570.2 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr15_+_55420950 | 0.57 |
ENSMUST00000170046.8
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr11_+_26337194 | 0.52 |
ENSMUST00000136830.2
ENSMUST00000109509.8 |
Fancl
|
Fanconi anemia, complementation group L |
| chr3_+_32583602 | 0.50 |
ENSMUST00000091257.11
|
Mfn1
|
mitofusin 1 |
| chr2_-_91067212 | 0.49 |
ENSMUST00000111352.8
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr6_-_87827993 | 0.46 |
ENSMUST00000204890.3
ENSMUST00000113617.3 ENSMUST00000113619.8 ENSMUST00000204653.3 ENSMUST00000032138.15 |
Cnbp
|
cellular nucleic acid binding protein |
| chr9_-_35028100 | 0.45 |
ENSMUST00000034537.8
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_-_93729650 | 0.45 |
ENSMUST00000027503.14
|
Dtymk
|
deoxythymidylate kinase |
| chr5_-_33809640 | 0.45 |
ENSMUST00000151081.2
ENSMUST00000139518.8 ENSMUST00000101354.10 |
Slbp
|
stem-loop binding protein |
| chr5_-_33809872 | 0.44 |
ENSMUST00000057551.14
|
Slbp
|
stem-loop binding protein |
| chr10_-_21036792 | 0.44 |
ENSMUST00000188495.8
|
Myb
|
myeloblastosis oncogene |
| chr17_+_34128455 | 0.43 |
ENSMUST00000173626.8
ENSMUST00000170075.9 |
Daxx
|
Fas death domain-associated protein |
| chr8_+_57964956 | 0.42 |
ENSMUST00000210871.2
|
Hmgb2
|
high mobility group box 2 |
| chr6_+_117893942 | 0.41 |
ENSMUST00000179478.8
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr7_-_128342158 | 0.39 |
ENSMUST00000119081.2
ENSMUST00000057557.14 |
Mcmbp
|
minichromosome maintenance complex binding protein |
| chr10_+_59538467 | 0.38 |
ENSMUST00000171409.8
|
Micu1
|
mitochondrial calcium uptake 1 |
| chr15_+_55420795 | 0.34 |
ENSMUST00000022998.14
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr2_-_91067312 | 0.33 |
ENSMUST00000028696.5
|
Ddb2
|
damage specific DNA binding protein 2 |
| chr17_+_34128382 | 0.32 |
ENSMUST00000173028.8
ENSMUST00000079421.15 |
Daxx
|
Fas death domain-associated protein |
| chr2_-_154411765 | 0.29 |
ENSMUST00000103145.11
|
E2f1
|
E2F transcription factor 1 |
| chr2_-_154411640 | 0.26 |
ENSMUST00000000894.6
|
E2f1
|
E2F transcription factor 1 |
| chr6_+_116241146 | 0.25 |
ENSMUST00000112900.9
ENSMUST00000036503.14 ENSMUST00000223495.2 |
Zfand4
|
zinc finger, AN1-type domain 4 |
| chr16_-_18630365 | 0.24 |
ENSMUST00000096990.10
|
Cdc45
|
cell division cycle 45 |
| chr6_+_29348068 | 0.24 |
ENSMUST00000173216.8
ENSMUST00000173694.5 ENSMUST00000172974.8 ENSMUST00000031779.17 ENSMUST00000090481.14 |
Calu
|
calumenin |
| chr11_+_21522221 | 0.24 |
ENSMUST00000131135.3
ENSMUST00000020568.10 |
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chr3_+_32583681 | 0.24 |
ENSMUST00000147350.8
|
Mfn1
|
mitofusin 1 |
| chr6_+_38528738 | 0.24 |
ENSMUST00000161227.8
|
Luc7l2
|
LUC7-like 2 (S. cerevisiae) |
| chr16_-_15455141 | 0.23 |
ENSMUST00000023353.4
|
Mcm4
|
minichromosome maintenance complex component 4 |
| chr6_+_108760025 | 0.20 |
ENSMUST00000032196.9
|
Arl8b
|
ADP-ribosylation factor-like 8B |
| chr16_+_18066825 | 0.19 |
ENSMUST00000100099.10
|
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chrX_-_69520778 | 0.18 |
ENSMUST00000101506.10
ENSMUST00000114630.3 |
Tmem185a
|
transmembrane protein 185A |
| chr16_+_18066730 | 0.17 |
ENSMUST00000115640.8
ENSMUST00000140206.8 |
Trmt2a
|
TRM2 tRNA methyltransferase 2A |
| chr10_-_128361731 | 0.14 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr17_-_35454729 | 0.14 |
ENSMUST00000048994.7
|
Nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor like 1 |
| chr5_+_75312939 | 0.13 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr1_-_132067343 | 0.12 |
ENSMUST00000112362.3
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr3_-_49711765 | 0.09 |
ENSMUST00000035931.13
|
Pcdh18
|
protocadherin 18 |
| chr1_-_172034251 | 0.09 |
ENSMUST00000155109.2
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr3_-_49711706 | 0.09 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr10_-_21036824 | 0.09 |
ENSMUST00000020158.9
|
Myb
|
myeloblastosis oncogene |
| chr12_-_12991828 | 0.07 |
ENSMUST00000043396.15
|
Mycn
|
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
| chr13_+_44884757 | 0.06 |
ENSMUST00000174086.2
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr19_+_4806859 | 0.05 |
ENSMUST00000237921.2
|
Rbm4b
|
RNA binding motif protein 4B |
| chr11_-_99549786 | 0.04 |
ENSMUST00000092695.5
|
Gm11563
|
predicted gene 11563 |
| chr8_-_107792264 | 0.02 |
ENSMUST00000034393.7
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr6_+_21215472 | 0.02 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr12_+_84498196 | 0.01 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr16_+_10652910 | 0.00 |
ENSMUST00000037913.9
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr10_+_42736539 | 0.00 |
ENSMUST00000157071.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chrX_+_48559327 | 0.00 |
ENSMUST00000114904.10
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chrX_-_167970196 | 0.00 |
ENSMUST00000066112.12
ENSMUST00000112118.8 ENSMUST00000112120.8 ENSMUST00000112119.8 |
Amelx
|
amelogenin, X-linked |
| chrX_+_48559432 | 0.00 |
ENSMUST00000042444.7
|
Arhgap36
|
Rho GTPase activating protein 36 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.5 | 1.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.4 | 3.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 1.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.3 | 1.6 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.3 | 1.6 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.3 | 3.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 5.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 0.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 | 0.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 | 2.9 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.2 | 0.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.7 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 1.2 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.4 | GO:0060762 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.6 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 1.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 1.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.8 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.8 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 3.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.7 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 1.2 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 1.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 2.3 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.5 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 6.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 1.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.4 | 2.3 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 1.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 1.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 0.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.3 | 0.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 0.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 3.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.7 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 3.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.7 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 1.6 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 2.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.5 | 3.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 1.0 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.3 | 2.3 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.3 | 1.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 0.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.3 | 0.8 | GO:0032142 | single guanine insertion binding(GO:0032142) |
| 0.2 | 1.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 2.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 4.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.0 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.7 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.1 | 0.4 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.1 | 1.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 1.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.5 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 2.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 1.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 2.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.0 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 5.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 2.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 5.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 2.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 3.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.8 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.7 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.1 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |