Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Ebf3
Z-value: 0.53
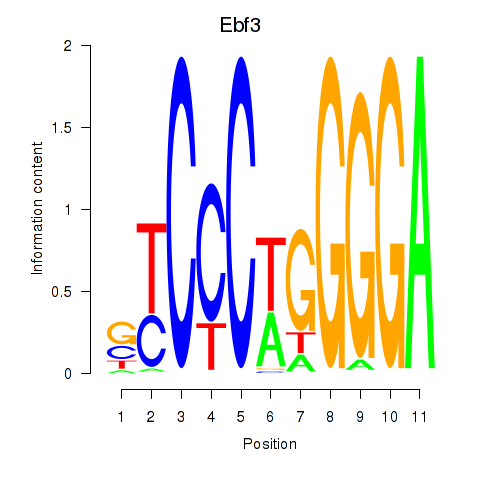
Transcription factors associated with Ebf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ebf3
|
ENSMUSG00000010476.15 | early B cell factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ebf3 | mm39_v1_chr7_-_136916123_136916174 | 0.81 | 1.0e-01 | Click! |
Activity profile of Ebf3 motif
Sorted Z-values of Ebf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102255999 | 0.59 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr1_+_173983199 | 0.21 |
ENSMUST00000213748.2
|
Olfr420
|
olfactory receptor 420 |
| chr8_+_13455080 | 0.21 |
ENSMUST00000033827.8
ENSMUST00000209909.2 |
Grk1
|
G protein-coupled receptor kinase 1 |
| chrX_+_7688528 | 0.19 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr7_-_102148780 | 0.19 |
ENSMUST00000216116.4
|
Olfr545
|
olfactory receptor 545 |
| chr7_-_68398989 | 0.19 |
ENSMUST00000048068.15
|
Arrdc4
|
arrestin domain containing 4 |
| chr14_-_40735710 | 0.18 |
ENSMUST00000143143.8
ENSMUST00000128236.2 ENSMUST00000022317.15 ENSMUST00000118466.8 |
Prxl2a
|
peroxiredoxin like 2A |
| chr2_-_27974889 | 0.17 |
ENSMUST00000028179.15
ENSMUST00000117486.8 ENSMUST00000135472.2 |
Fcnb
|
ficolin B |
| chr17_+_87270504 | 0.16 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr11_-_95733235 | 0.16 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr16_+_13721016 | 0.16 |
ENSMUST00000128757.8
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr7_+_126380655 | 0.15 |
ENSMUST00000172352.8
ENSMUST00000094037.5 |
Tbx6
|
T-box 6 |
| chr14_-_69740670 | 0.15 |
ENSMUST00000180059.3
ENSMUST00000179116.3 |
Gm16867
|
predicted gene, 16867 |
| chr11_+_117198958 | 0.14 |
ENSMUST00000153668.8
|
Septin9
|
septin 9 |
| chr7_+_123582021 | 0.13 |
ENSMUST00000106437.2
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr7_-_44465043 | 0.13 |
ENSMUST00000107893.9
|
Atf5
|
activating transcription factor 5 |
| chr5_+_123280250 | 0.12 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr10_+_58649181 | 0.12 |
ENSMUST00000135526.9
ENSMUST00000153031.2 |
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr13_+_30844025 | 0.11 |
ENSMUST00000110310.9
ENSMUST00000095914.7 |
Dusp22
|
dual specificity phosphatase 22 |
| chr7_-_139941566 | 0.11 |
ENSMUST00000215023.2
ENSMUST00000216027.2 ENSMUST00000210932.3 ENSMUST00000211031.3 |
Olfr60
|
olfactory receptor 60 |
| chr12_-_113912416 | 0.11 |
ENSMUST00000103464.3
|
Ighv4-1
|
immunoglobulin heavy variable 4-1 |
| chrX_+_10351360 | 0.11 |
ENSMUST00000076354.13
ENSMUST00000115526.2 |
Tspan7
|
tetraspanin 7 |
| chr13_+_55097200 | 0.11 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr10_-_12490424 | 0.11 |
ENSMUST00000217994.2
|
Utrn
|
utrophin |
| chr2_+_111144362 | 0.11 |
ENSMUST00000219291.2
|
Olfr1280
|
olfactory receptor 1280 |
| chr7_+_4693603 | 0.11 |
ENSMUST00000120836.8
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr7_+_4693759 | 0.11 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr13_-_49462694 | 0.11 |
ENSMUST00000110087.9
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr6_-_42453259 | 0.11 |
ENSMUST00000204324.3
ENSMUST00000203396.3 |
Olfr457
|
olfactory receptor 457 |
| chr19_-_47452557 | 0.10 |
ENSMUST00000111800.4
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr19_+_8828132 | 0.10 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr2_+_93017887 | 0.10 |
ENSMUST00000150462.8
ENSMUST00000111266.8 |
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr9_+_38374440 | 0.10 |
ENSMUST00000216724.3
|
Olfr904
|
olfactory receptor 904 |
| chr8_-_46452896 | 0.10 |
ENSMUST00000053558.10
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr7_-_68398917 | 0.10 |
ENSMUST00000118110.3
|
Arrdc4
|
arrestin domain containing 4 |
| chr16_+_24266829 | 0.10 |
ENSMUST00000078988.10
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_-_116128903 | 0.09 |
ENSMUST00000037007.4
|
Evpl
|
envoplakin |
| chr5_-_147831610 | 0.09 |
ENSMUST00000118527.8
ENSMUST00000031655.4 ENSMUST00000138244.2 |
Slc46a3
|
solute carrier family 46, member 3 |
| chr5_+_23992689 | 0.09 |
ENSMUST00000120869.6
ENSMUST00000030852.13 ENSMUST00000117783.8 ENSMUST00000115113.3 |
Rint1
|
RAD50 interactor 1 |
| chr3_-_89294430 | 0.09 |
ENSMUST00000107433.8
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr11_+_95733489 | 0.09 |
ENSMUST00000100532.10
ENSMUST00000036088.11 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr10_-_60055082 | 0.09 |
ENSMUST00000135158.9
|
Chst3
|
carbohydrate sulfotransferase 3 |
| chr9_+_31191820 | 0.09 |
ENSMUST00000117389.8
ENSMUST00000215499.2 |
Prdm10
|
PR domain containing 10 |
| chr4_-_133484080 | 0.09 |
ENSMUST00000008024.7
|
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr13_+_113130374 | 0.09 |
ENSMUST00000092089.6
|
Mcidas
|
multiciliate differentiation and DNA synthesis associated cell cycle protein |
| chr16_+_32427789 | 0.09 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr17_+_44499451 | 0.09 |
ENSMUST00000024755.7
|
Clic5
|
chloride intracellular channel 5 |
| chr5_-_137312424 | 0.09 |
ENSMUST00000199121.2
|
Trip6
|
thyroid hormone receptor interactor 6 |
| chr4_+_129030710 | 0.08 |
ENSMUST00000102600.4
|
Fndc5
|
fibronectin type III domain containing 5 |
| chr10_+_79986988 | 0.08 |
ENSMUST00000146516.8
ENSMUST00000144526.2 |
Midn
|
midnolin |
| chr19_+_13385213 | 0.08 |
ENSMUST00000216910.3
|
Olfr1469
|
olfactory receptor 1469 |
| chr18_+_36481706 | 0.08 |
ENSMUST00000235864.2
ENSMUST00000050584.10 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr7_+_24596806 | 0.08 |
ENSMUST00000003469.8
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr7_-_102507962 | 0.08 |
ENSMUST00000213481.2
ENSMUST00000209952.2 |
Olfr566
|
olfactory receptor 566 |
| chr1_+_176642226 | 0.08 |
ENSMUST00000056773.15
ENSMUST00000027785.15 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr15_+_8198506 | 0.08 |
ENSMUST00000110617.2
|
Cplane1
|
ciliogenesis and planar polarity effector 1 |
| chr11_-_69771797 | 0.08 |
ENSMUST00000238978.2
|
Kctd11
|
potassium channel tetramerisation domain containing 11 |
| chr11_+_98818640 | 0.08 |
ENSMUST00000107474.8
|
Rara
|
retinoic acid receptor, alpha |
| chr17_-_74017410 | 0.08 |
ENSMUST00000112591.3
ENSMUST00000024858.12 |
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr15_-_66684442 | 0.08 |
ENSMUST00000100572.10
|
Sla
|
src-like adaptor |
| chr13_-_93810808 | 0.08 |
ENSMUST00000015941.8
|
Bhmt2
|
betaine-homocysteine methyltransferase 2 |
| chr2_-_28730286 | 0.07 |
ENSMUST00000037117.6
ENSMUST00000171404.8 |
Gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr15_+_86098660 | 0.07 |
ENSMUST00000063414.9
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chrX_-_7834057 | 0.07 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr19_-_10847121 | 0.07 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr7_+_101027390 | 0.07 |
ENSMUST00000084895.12
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr15_-_71906051 | 0.07 |
ENSMUST00000159993.8
|
Col22a1
|
collagen, type XXII, alpha 1 |
| chr16_+_32427738 | 0.07 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr6_-_116650751 | 0.07 |
ENSMUST00000204576.2
ENSMUST00000203029.3 ENSMUST00000035842.7 |
Rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr4_+_83443777 | 0.07 |
ENSMUST00000053414.13
ENSMUST00000231339.2 ENSMUST00000126429.8 |
Ccdc171
|
coiled-coil domain containing 171 |
| chr4_-_155430153 | 0.07 |
ENSMUST00000103178.11
|
Prkcz
|
protein kinase C, zeta |
| chr6_-_124441731 | 0.06 |
ENSMUST00000008297.5
|
Clstn3
|
calsyntenin 3 |
| chr7_+_79676095 | 0.06 |
ENSMUST00000206039.2
|
Zfp710
|
zinc finger protein 710 |
| chr11_-_99045894 | 0.06 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr8_+_107141948 | 0.06 |
ENSMUST00000034382.8
ENSMUST00000212606.2 |
Zfp90
|
zinc finger protein 90 |
| chr6_-_69415741 | 0.06 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr2_+_34723019 | 0.06 |
ENSMUST00000201786.4
|
Gm34653
|
predicted gene, 34653 |
| chr6_+_142359099 | 0.06 |
ENSMUST00000126521.9
ENSMUST00000211094.2 |
Spx
|
spexin hormone |
| chr8_-_95644639 | 0.06 |
ENSMUST00000077955.6
|
Ccdc102a
|
coiled-coil domain containing 102A |
| chr12_-_31763859 | 0.06 |
ENSMUST00000057783.6
ENSMUST00000236002.2 ENSMUST00000174480.3 ENSMUST00000176710.2 |
Gpr22
|
G protein-coupled receptor 22 |
| chr8_+_107142031 | 0.06 |
ENSMUST00000212874.2
|
Zfp90
|
zinc finger protein 90 |
| chr4_-_47474283 | 0.06 |
ENSMUST00000044148.3
|
Alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr11_-_59397352 | 0.06 |
ENSMUST00000013262.15
ENSMUST00000101150.9 |
Zkscan17
|
zinc finger with KRAB and SCAN domains 17 |
| chr7_+_27879650 | 0.06 |
ENSMUST00000172467.8
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr15_+_76594810 | 0.06 |
ENSMUST00000136840.8
ENSMUST00000127208.8 ENSMUST00000036423.15 ENSMUST00000137649.8 ENSMUST00000155225.2 ENSMUST00000155735.2 |
Lrrc14
|
leucine rich repeat containing 14 |
| chr5_-_144698443 | 0.06 |
ENSMUST00000061446.8
|
Tmem130
|
transmembrane protein 130 |
| chrX_+_47197789 | 0.06 |
ENSMUST00000114998.8
ENSMUST00000115000.4 |
Xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr5_-_147337162 | 0.06 |
ENSMUST00000049324.13
|
Flt3
|
FMS-like tyrosine kinase 3 |
| chr6_-_124519240 | 0.06 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr7_+_104210108 | 0.05 |
ENSMUST00000219111.2
ENSMUST00000215410.2 ENSMUST00000216131.2 |
Olfr652
|
olfactory receptor 652 |
| chr14_-_70864666 | 0.05 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr11_-_97635484 | 0.05 |
ENSMUST00000018691.9
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chrX_+_73352694 | 0.05 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr11_-_77498636 | 0.05 |
ENSMUST00000058496.8
|
Taok1
|
TAO kinase 1 |
| chr1_-_131441962 | 0.05 |
ENSMUST00000185445.3
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr11_-_114851243 | 0.05 |
ENSMUST00000092466.13
ENSMUST00000061637.4 |
Cd300c
|
CD300C molecule |
| chr13_-_21826688 | 0.05 |
ENSMUST00000205631.3
|
Olfr11
|
olfactory receptor 11 |
| chr16_-_13720915 | 0.05 |
ENSMUST00000115803.9
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr11_-_113956996 | 0.05 |
ENSMUST00000041627.14
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr16_-_13720949 | 0.05 |
ENSMUST00000023361.12
ENSMUST00000115802.2 |
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr2_-_37007795 | 0.05 |
ENSMUST00000213817.2
ENSMUST00000215927.2 |
Olfr362
|
olfactory receptor 362 |
| chr5_-_92110927 | 0.05 |
ENSMUST00000031345.15
|
Rchy1
|
ring finger and CHY zinc finger domain containing 1 |
| chr2_+_59442378 | 0.05 |
ENSMUST00000112568.8
ENSMUST00000037526.11 |
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr18_+_31767369 | 0.05 |
ENSMUST00000235017.2
ENSMUST00000178164.8 ENSMUST00000025109.8 |
Sap130
|
Sin3A associated protein |
| chr5_+_137627376 | 0.05 |
ENSMUST00000031734.16
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr16_+_32734464 | 0.05 |
ENSMUST00000023491.13
ENSMUST00000170899.8 ENSMUST00000170201.8 ENSMUST00000165616.8 ENSMUST00000135193.9 |
Lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr7_-_3680530 | 0.05 |
ENSMUST00000038743.15
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr5_-_97259447 | 0.05 |
ENSMUST00000069453.9
ENSMUST00000112968.2 |
Paqr3
|
progestin and adipoQ receptor family member III |
| chr11_-_69728560 | 0.05 |
ENSMUST00000108634.9
|
Nlgn2
|
neuroligin 2 |
| chr6_+_68279392 | 0.05 |
ENSMUST00000103322.3
|
Igkv2-109
|
immunoglobulin kappa variable 2-109 |
| chr17_+_87270707 | 0.05 |
ENSMUST00000139344.2
|
Rhoq
|
ras homolog family member Q |
| chr1_-_63153414 | 0.05 |
ENSMUST00000153992.2
ENSMUST00000165066.8 ENSMUST00000172416.8 ENSMUST00000137511.8 |
Ino80d
|
INO80 complex subunit D |
| chr4_+_117692583 | 0.05 |
ENSMUST00000169885.8
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr1_-_24139387 | 0.04 |
ENSMUST00000027337.15
|
Fam135a
|
family with sequence similarity 135, member A |
| chr7_+_141996067 | 0.04 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr4_-_148215135 | 0.04 |
ENSMUST00000030862.5
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr17_-_34340918 | 0.04 |
ENSMUST00000151986.2
|
Brd2
|
bromodomain containing 2 |
| chr13_+_42205491 | 0.04 |
ENSMUST00000060148.6
|
Hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chrX_-_100463810 | 0.04 |
ENSMUST00000118092.8
ENSMUST00000119699.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr17_+_47983587 | 0.04 |
ENSMUST00000152724.2
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr7_+_102954855 | 0.04 |
ENSMUST00000214577.2
|
Olfr596
|
olfactory receptor 596 |
| chr17_+_46989695 | 0.04 |
ENSMUST00000233032.2
|
Mea1
|
male enhanced antigen 1 |
| chr1_-_165462020 | 0.04 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chrX_+_158242121 | 0.04 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr9_+_108685555 | 0.04 |
ENSMUST00000035218.9
ENSMUST00000195323.2 ENSMUST00000194819.2 |
Nckipsd
|
NCK interacting protein with SH3 domain |
| chr11_+_21041291 | 0.04 |
ENSMUST00000093290.12
|
Peli1
|
pellino 1 |
| chr3_+_97066065 | 0.04 |
ENSMUST00000090759.5
|
Acp6
|
acid phosphatase 6, lysophosphatidic |
| chr2_+_88479038 | 0.04 |
ENSMUST00000120518.4
|
Olfr1192-ps1
|
olfactory receptor 1192, pseudogene 1 |
| chr3_+_102642272 | 0.04 |
ENSMUST00000196611.5
|
Tspan2
|
tetraspanin 2 |
| chr11_+_104122216 | 0.04 |
ENSMUST00000106992.10
|
Mapt
|
microtubule-associated protein tau |
| chr15_+_99600475 | 0.04 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr7_+_27307422 | 0.04 |
ENSMUST00000142365.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr7_+_144468837 | 0.04 |
ENSMUST00000128057.8
ENSMUST00000033388.12 ENSMUST00000141737.2 ENSMUST00000105895.2 |
LTO1
|
ABCE maturation factor |
| chr2_-_90410922 | 0.04 |
ENSMUST00000168621.3
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr9_-_44231526 | 0.04 |
ENSMUST00000214602.2
ENSMUST00000065080.10 |
C2cd2l
|
C2 calcium-dependent domain containing 2-like |
| chr5_-_74229021 | 0.04 |
ENSMUST00000119154.8
ENSMUST00000068058.14 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr1_-_57008986 | 0.04 |
ENSMUST00000176759.2
ENSMUST00000177424.2 |
Satb2
|
special AT-rich sequence binding protein 2 |
| chr11_+_92989229 | 0.04 |
ENSMUST00000107859.8
ENSMUST00000107861.8 ENSMUST00000042943.13 ENSMUST00000107858.9 |
Car10
|
carbonic anhydrase 10 |
| chr1_-_173195236 | 0.04 |
ENSMUST00000005470.5
ENSMUST00000111220.8 |
Cadm3
|
cell adhesion molecule 3 |
| chr17_+_84013575 | 0.04 |
ENSMUST00000112350.8
ENSMUST00000112349.9 ENSMUST00000112352.10 ENSMUST00000067826.15 |
Mta3
|
metastasis associated 3 |
| chr4_-_131988832 | 0.04 |
ENSMUST00000105965.8
|
Gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr1_+_89382491 | 0.04 |
ENSMUST00000027521.15
ENSMUST00000190096.7 |
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr2_+_30023758 | 0.04 |
ENSMUST00000044556.12
|
Tbc1d13
|
TBC1 domain family, member 13 |
| chr4_+_119590971 | 0.03 |
ENSMUST00000084306.5
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr1_-_24139263 | 0.03 |
ENSMUST00000187369.7
ENSMUST00000187752.7 ENSMUST00000186999.7 |
Fam135a
|
family with sequence similarity 135, member A |
| chr14_-_65187287 | 0.03 |
ENSMUST00000067843.10
ENSMUST00000176489.8 ENSMUST00000175905.8 ENSMUST00000022544.14 ENSMUST00000175744.8 ENSMUST00000176128.8 |
Hmbox1
|
homeobox containing 1 |
| chr8_-_72154405 | 0.03 |
ENSMUST00000034260.9
|
B3gnt3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr7_+_140181182 | 0.03 |
ENSMUST00000214180.2
ENSMUST00000211771.2 |
Olfr46
|
olfactory receptor 46 |
| chr11_+_16901370 | 0.03 |
ENSMUST00000109635.2
ENSMUST00000061327.2 |
Fbxo48
|
F-box protein 48 |
| chr1_-_74788013 | 0.03 |
ENSMUST00000188073.7
|
Prkag3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit |
| chr4_+_152181155 | 0.03 |
ENSMUST00000105661.10
ENSMUST00000084115.4 |
Plekhg5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr14_-_60324265 | 0.03 |
ENSMUST00000080368.13
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr3_+_102642516 | 0.03 |
ENSMUST00000119902.6
|
Tspan2
|
tetraspanin 2 |
| chr1_-_93406091 | 0.03 |
ENSMUST00000188165.2
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr13_-_96028412 | 0.03 |
ENSMUST00000068603.8
|
Iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr6_-_116650728 | 0.03 |
ENSMUST00000204203.2
|
Rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr2_+_26209755 | 0.03 |
ENSMUST00000066889.13
|
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr3_-_53370621 | 0.03 |
ENSMUST00000056749.14
|
Nhlrc3
|
NHL repeat containing 3 |
| chrX_-_161426542 | 0.03 |
ENSMUST00000101102.2
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr2_+_87436556 | 0.03 |
ENSMUST00000213103.3
ENSMUST00000216580.2 |
Olfr1130
|
olfactory receptor 1130 |
| chr6_-_69204417 | 0.03 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr7_-_126807581 | 0.03 |
ENSMUST00000120705.3
|
Tbc1d10b
|
TBC1 domain family, member 10b |
| chr13_+_30843937 | 0.03 |
ENSMUST00000091672.13
|
Dusp22
|
dual specificity phosphatase 22 |
| chr9_+_103182352 | 0.03 |
ENSMUST00000035164.10
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chr6_-_69245427 | 0.03 |
ENSMUST00000103348.3
|
Igkv4-70
|
immunoglobulin kappa chain variable 4-70 |
| chr12_-_113823290 | 0.03 |
ENSMUST00000103459.5
|
Ighv5-17
|
immunoglobulin heavy variable 5-17 |
| chr11_-_72686853 | 0.03 |
ENSMUST00000156294.8
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr7_+_101043568 | 0.03 |
ENSMUST00000098243.4
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_+_149561835 | 0.03 |
ENSMUST00000210722.3
|
Tmem274
|
transmembrame protein 274 |
| chr15_-_76084776 | 0.03 |
ENSMUST00000169108.8
ENSMUST00000170728.8 |
Plec
|
plectin |
| chr15_+_81511486 | 0.03 |
ENSMUST00000206833.2
|
Ep300
|
E1A binding protein p300 |
| chr10_-_67748461 | 0.03 |
ENSMUST00000064656.8
|
Zfp365
|
zinc finger protein 365 |
| chr9_-_110572721 | 0.03 |
ENSMUST00000166716.8
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr5_+_145104011 | 0.03 |
ENSMUST00000160629.8
ENSMUST00000070487.12 ENSMUST00000160422.8 ENSMUST00000162244.8 |
Cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr7_+_140261864 | 0.03 |
ENSMUST00000214637.3
|
Olfr45
|
olfactory receptor 45 |
| chr6_+_22288220 | 0.03 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr5_+_150042092 | 0.02 |
ENSMUST00000200960.4
ENSMUST00000202530.4 |
Fry
|
FRY microtubule binding protein |
| chr5_-_123662175 | 0.02 |
ENSMUST00000200247.5
ENSMUST00000111586.8 ENSMUST00000031385.7 ENSMUST00000145152.8 ENSMUST00000111587.10 |
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr11_-_29775649 | 0.02 |
ENSMUST00000104962.2
|
Fem1al
|
fem-1 homolog A like |
| chr14_-_70866385 | 0.02 |
ENSMUST00000228824.2
|
Dmtn
|
dematin actin binding protein |
| chr1_+_74582044 | 0.02 |
ENSMUST00000113749.8
ENSMUST00000067916.13 ENSMUST00000113747.8 ENSMUST00000113750.8 |
Plcd4
|
phospholipase C, delta 4 |
| chr9_+_89791943 | 0.02 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr17_-_48758538 | 0.02 |
ENSMUST00000024794.12
|
Tspo2
|
translocator protein 2 |
| chr12_-_113625906 | 0.02 |
ENSMUST00000103448.3
|
Ighv5-9
|
immunoglobulin heavy variable 5-9 |
| chr10_+_57508077 | 0.02 |
ENSMUST00000095668.10
ENSMUST00000075992.12 |
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr6_-_32565127 | 0.02 |
ENSMUST00000115096.4
|
Plxna4
|
plexin A4 |
| chr3_+_115681788 | 0.02 |
ENSMUST00000196804.5
ENSMUST00000106505.8 ENSMUST00000043342.10 |
Dph5
|
diphthamide biosynthesis 5 |
| chr1_+_156386414 | 0.02 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr11_+_96820091 | 0.02 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr14_+_56905698 | 0.02 |
ENSMUST00000116468.2
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr7_-_44741622 | 0.02 |
ENSMUST00000210469.2
ENSMUST00000211352.2 ENSMUST00000019683.11 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr4_-_149760488 | 0.02 |
ENSMUST00000118704.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr11_+_97692738 | 0.02 |
ENSMUST00000127033.9
|
Lasp1
|
LIM and SH3 protein 1 |
| chr14_+_21881794 | 0.02 |
ENSMUST00000152562.8
|
Vdac2
|
voltage-dependent anion channel 2 |
| chr1_+_183078851 | 0.02 |
ENSMUST00000193625.2
|
Aida
|
axin interactor, dorsalization associated |
| chr15_+_102011352 | 0.02 |
ENSMUST00000169627.9
|
Tns2
|
tensin 2 |
| chr12_-_113860566 | 0.02 |
ENSMUST00000103474.5
|
Ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr3_+_94745009 | 0.02 |
ENSMUST00000107266.8
ENSMUST00000042402.12 ENSMUST00000107269.2 |
Pogz
|
pogo transposable element with ZNF domain |
| chr17_-_57181420 | 0.02 |
ENSMUST00000043062.5
|
Acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr12_-_113561594 | 0.02 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ebf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0097021 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.0 | 0.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0036145 | dendritic cell homeostasis(GO:0036145) |
| 0.0 | 0.1 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.0 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |