Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
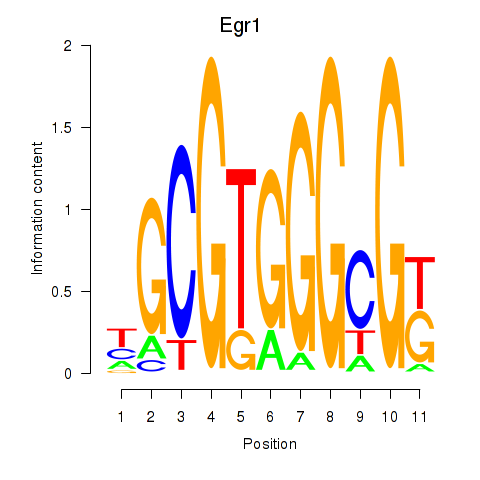
Results for Egr1
Z-value: 0.74
Transcription factors associated with Egr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr1
|
ENSMUSG00000038418.8 | early growth response 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr1 | mm39_v1_chr18_+_34994253_34994268 | 0.59 | 3.0e-01 | Click! |
Activity profile of Egr1 motif
Sorted Z-values of Egr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_21938258 | 0.65 |
ENSMUST00000091709.3
|
H2bc15
|
H2B clustered histone 15 |
| chrX_-_20787150 | 0.49 |
ENSMUST00000081893.7
ENSMUST00000115345.8 |
Syn1
|
synapsin I |
| chr13_+_23715220 | 0.47 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr6_+_122803624 | 0.45 |
ENSMUST00000203075.2
|
Foxj2
|
forkhead box J2 |
| chr2_+_117942357 | 0.42 |
ENSMUST00000039559.9
|
Thbs1
|
thrombospondin 1 |
| chr4_+_42949814 | 0.40 |
ENSMUST00000037872.10
ENSMUST00000098112.9 |
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr8_+_73072877 | 0.39 |
ENSMUST00000067912.8
|
Klf2
|
Kruppel-like factor 2 (lung) |
| chr6_-_52211882 | 0.37 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr5_-_107873883 | 0.29 |
ENSMUST00000159263.3
|
Gfi1
|
growth factor independent 1 transcription repressor |
| chr2_-_34262012 | 0.28 |
ENSMUST00000113132.9
ENSMUST00000040638.15 |
Pbx3
|
pre B cell leukemia homeobox 3 |
| chr11_+_118913788 | 0.24 |
ENSMUST00000026662.8
|
Cbx2
|
chromobox 2 |
| chr10_+_79977291 | 0.24 |
ENSMUST00000105367.8
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr1_-_36978602 | 0.23 |
ENSMUST00000027290.12
|
Tmem131
|
transmembrane protein 131 |
| chr2_+_32425327 | 0.23 |
ENSMUST00000133512.2
ENSMUST00000048375.6 |
Fam102a
|
family with sequence similarity 102, member A |
| chr11_+_98851238 | 0.22 |
ENSMUST00000107473.3
|
Rara
|
retinoic acid receptor, alpha |
| chr5_-_142891686 | 0.21 |
ENSMUST00000106216.3
|
Actb
|
actin, beta |
| chr14_-_33169099 | 0.21 |
ENSMUST00000111944.10
ENSMUST00000022504.12 ENSMUST00000111945.9 |
Mapk8
|
mitogen-activated protein kinase 8 |
| chr1_-_86286690 | 0.21 |
ENSMUST00000185785.2
|
Ncl
|
nucleolin |
| chr14_+_24540745 | 0.21 |
ENSMUST00000112384.10
|
Rps24
|
ribosomal protein S24 |
| chr6_-_124894902 | 0.21 |
ENSMUST00000032216.7
|
Ptms
|
parathymosin |
| chr19_+_37423198 | 0.20 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr3_-_131196213 | 0.20 |
ENSMUST00000197057.2
|
Sgms2
|
sphingomyelin synthase 2 |
| chr1_+_39940189 | 0.19 |
ENSMUST00000191761.6
ENSMUST00000193682.6 ENSMUST00000195860.6 ENSMUST00000195259.6 ENSMUST00000195636.6 ENSMUST00000192509.6 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr17_-_35340189 | 0.18 |
ENSMUST00000025246.13
ENSMUST00000173114.8 |
Csnk2b
|
casein kinase 2, beta polypeptide |
| chr14_+_24540815 | 0.18 |
ENSMUST00000224568.2
|
Rps24
|
ribosomal protein S24 |
| chr8_-_33374282 | 0.17 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chr5_-_137015683 | 0.17 |
ENSMUST00000034953.14
ENSMUST00000085941.12 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr5_+_143608194 | 0.17 |
ENSMUST00000116456.10
|
Cyth3
|
cytohesin 3 |
| chr7_+_26958150 | 0.17 |
ENSMUST00000079258.7
|
Numbl
|
numb-like |
| chr14_+_24540777 | 0.16 |
ENSMUST00000169826.3
ENSMUST00000225023.2 ENSMUST00000223999.2 |
Rps24
|
ribosomal protein S24 |
| chr19_+_4264470 | 0.16 |
ENSMUST00000237171.2
|
Gm45928
|
predicted gene, 45928 |
| chr17_-_35340006 | 0.15 |
ENSMUST00000174306.8
ENSMUST00000174024.8 |
Csnk2b
|
casein kinase 2, beta polypeptide |
| chr7_-_45071897 | 0.15 |
ENSMUST00000210271.2
|
Ruvbl2
|
RuvB-like protein 2 |
| chr3_+_146205864 | 0.15 |
ENSMUST00000119130.2
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr2_-_27365612 | 0.15 |
ENSMUST00000147736.2
|
Brd3
|
bromodomain containing 3 |
| chr4_+_43669266 | 0.14 |
ENSMUST00000107864.8
|
Tmem8b
|
transmembrane protein 8B |
| chr4_+_42917228 | 0.14 |
ENSMUST00000107976.9
ENSMUST00000069184.9 |
Phf24
|
PHD finger protein 24 |
| chr11_-_106679671 | 0.14 |
ENSMUST00000123339.2
|
Ddx5
|
DEAD box helicase 5 |
| chr18_-_20879461 | 0.14 |
ENSMUST00000070080.6
|
B4galt6
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6 |
| chr4_-_133856025 | 0.14 |
ENSMUST00000105879.2
ENSMUST00000030651.9 |
Sh3bgrl3
|
SH3 domain binding glutamic acid-rich protein-like 3 |
| chr11_-_4696778 | 0.14 |
ENSMUST00000009219.3
|
Cabp7
|
calcium binding protein 7 |
| chr17_-_32503060 | 0.14 |
ENSMUST00000003726.16
ENSMUST00000121285.8 ENSMUST00000120276.9 |
Brd4
|
bromodomain containing 4 |
| chrX_+_158410229 | 0.13 |
ENSMUST00000112456.9
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_+_80191692 | 0.13 |
ENSMUST00000017836.8
|
Rhbdl3
|
rhomboid like 3 |
| chr10_-_127502467 | 0.13 |
ENSMUST00000099157.4
|
Nab2
|
Ngfi-A binding protein 2 |
| chr9_+_118755521 | 0.13 |
ENSMUST00000073109.12
|
Ctdspl
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr14_+_76082736 | 0.13 |
ENSMUST00000142061.3
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr16_+_93680783 | 0.13 |
ENSMUST00000023666.11
ENSMUST00000117099.8 |
Chaf1b
|
chromatin assembly factor 1, subunit B (p60) |
| chr16_+_10363222 | 0.13 |
ENSMUST00000155633.8
ENSMUST00000066345.15 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr14_+_34542053 | 0.13 |
ENSMUST00000043349.7
|
Grid1
|
glutamate receptor, ionotropic, delta 1 |
| chr16_+_10363203 | 0.12 |
ENSMUST00000115824.10
|
Clec16a
|
C-type lectin domain family 16, member A |
| chr6_+_29468067 | 0.12 |
ENSMUST00000143101.4
ENSMUST00000149646.3 |
Atp6v1f
|
ATPase, H+ transporting, lysosomal V1 subunit F |
| chr12_+_108145997 | 0.12 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr16_-_18066591 | 0.12 |
ENSMUST00000115645.10
|
Ranbp1
|
RAN binding protein 1 |
| chr10_-_127502541 | 0.12 |
ENSMUST00000026469.9
|
Nab2
|
Ngfi-A binding protein 2 |
| chr2_+_164328375 | 0.12 |
ENSMUST00000069385.15
ENSMUST00000143690.8 |
Dbndd2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr5_-_139115417 | 0.12 |
ENSMUST00000026973.14
|
Prkar1b
|
protein kinase, cAMP dependent regulatory, type I beta |
| chr15_-_66158445 | 0.11 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chr7_-_126102579 | 0.11 |
ENSMUST00000040202.15
|
Atxn2l
|
ataxin 2-like |
| chr1_+_39940043 | 0.11 |
ENSMUST00000168431.7
ENSMUST00000163854.9 |
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr4_+_40723083 | 0.11 |
ENSMUST00000149794.2
|
Dnaja1
|
DnaJ heat shock protein family (Hsp40) member A1 |
| chr16_+_33504740 | 0.11 |
ENSMUST00000232568.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr3_+_146205562 | 0.11 |
ENSMUST00000090031.12
ENSMUST00000118280.2 |
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr7_-_126102470 | 0.11 |
ENSMUST00000206577.2
|
Atxn2l
|
ataxin 2-like |
| chr14_+_76082533 | 0.11 |
ENSMUST00000110894.9
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr1_+_166828982 | 0.11 |
ENSMUST00000165874.8
ENSMUST00000190081.7 |
Fam78b
|
family with sequence similarity 78, member B |
| chr8_-_81466126 | 0.11 |
ENSMUST00000043359.9
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr5_+_135717890 | 0.11 |
ENSMUST00000005651.13
ENSMUST00000122113.8 |
Por
|
P450 (cytochrome) oxidoreductase |
| chr2_+_27566452 | 0.10 |
ENSMUST00000129514.8
|
Rxra
|
retinoid X receptor alpha |
| chr11_+_115078299 | 0.10 |
ENSMUST00000100235.9
ENSMUST00000061450.7 |
Tmem104
|
transmembrane protein 104 |
| chr9_+_62746055 | 0.10 |
ENSMUST00000034776.13
|
Cln6
|
ceroid-lipofuscinosis, neuronal 6 |
| chr9_+_59658156 | 0.10 |
ENSMUST00000136740.8
ENSMUST00000135298.8 ENSMUST00000128341.2 |
Myo9a
|
myosin IXa |
| chr9_-_102231884 | 0.10 |
ENSMUST00000035129.14
ENSMUST00000085169.12 ENSMUST00000149800.3 |
Ephb1
|
Eph receptor B1 |
| chr7_+_127566629 | 0.10 |
ENSMUST00000106251.10
ENSMUST00000077609.12 ENSMUST00000121616.9 |
Fus
|
fused in sarcoma |
| chr15_+_78819378 | 0.10 |
ENSMUST00000145157.2
ENSMUST00000123013.2 |
Nol12
|
nucleolar protein 12 |
| chr4_-_93223746 | 0.10 |
ENSMUST00000066774.6
|
Tusc1
|
tumor suppressor candidate 1 |
| chr2_-_167030706 | 0.10 |
ENSMUST00000207917.2
|
Kcnb1
|
potassium voltage gated channel, Shab-related subfamily, member 1 |
| chr17_+_64907697 | 0.10 |
ENSMUST00000086723.10
|
Man2a1
|
mannosidase 2, alpha 1 |
| chr6_-_124733441 | 0.10 |
ENSMUST00000088357.12
|
Atn1
|
atrophin 1 |
| chr18_-_80243812 | 0.10 |
ENSMUST00000025462.7
|
Rbfa
|
ribosome binding factor A |
| chr7_+_101619897 | 0.10 |
ENSMUST00000211272.2
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr19_+_6413703 | 0.10 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr11_-_102298281 | 0.10 |
ENSMUST00000107098.8
ENSMUST00000018821.9 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr8_-_123405392 | 0.10 |
ENSMUST00000134045.2
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr19_+_53588808 | 0.10 |
ENSMUST00000025930.10
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr9_-_56325344 | 0.10 |
ENSMUST00000061552.15
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr5_+_137786031 | 0.09 |
ENSMUST00000035852.14
|
Zcwpw1
|
zinc finger, CW type with PWWP domain 1 |
| chr15_+_78784043 | 0.09 |
ENSMUST00000001226.11
|
Sh3bp1
|
SH3-domain binding protein 1 |
| chr9_-_44145280 | 0.09 |
ENSMUST00000205968.2
ENSMUST00000206147.2 ENSMUST00000037644.8 |
Cbl
|
Casitas B-lineage lymphoma |
| chr17_-_35420972 | 0.09 |
ENSMUST00000167924.2
ENSMUST00000025263.15 |
Tnf
|
tumor necrosis factor |
| chr19_+_5618029 | 0.09 |
ENSMUST00000235575.2
ENSMUST00000235542.2 |
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr5_-_137016355 | 0.09 |
ENSMUST00000137272.2
ENSMUST00000111090.9 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr19_+_6413840 | 0.09 |
ENSMUST00000113488.8
ENSMUST00000113487.8 |
Sf1
|
splicing factor 1 |
| chr11_-_104333059 | 0.09 |
ENSMUST00000106977.8
ENSMUST00000106972.8 |
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr1_-_156766351 | 0.09 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr7_-_126303351 | 0.09 |
ENSMUST00000106364.8
|
Coro1a
|
coronin, actin binding protein 1A |
| chr7_-_126303947 | 0.09 |
ENSMUST00000032949.14
|
Coro1a
|
coronin, actin binding protein 1A |
| chr9_+_44245981 | 0.08 |
ENSMUST00000052686.4
|
H2ax
|
H2A.X variant histone |
| chr19_+_23736205 | 0.08 |
ENSMUST00000025830.9
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr4_+_118965992 | 0.08 |
ENSMUST00000134105.8
ENSMUST00000144329.8 ENSMUST00000208090.2 |
Slc2a1
|
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr5_-_34093678 | 0.08 |
ENSMUST00000030993.8
|
Nelfa
|
negative elongation factor complex member A, Whsc2 |
| chr4_+_86666764 | 0.08 |
ENSMUST00000045512.15
ENSMUST00000082026.14 |
Dennd4c
|
DENN/MADD domain containing 4C |
| chrX_+_158410528 | 0.08 |
ENSMUST00000073094.10
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr19_+_5618096 | 0.08 |
ENSMUST00000096318.4
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr7_+_3648264 | 0.08 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr7_-_80338600 | 0.08 |
ENSMUST00000122255.8
|
Crtc3
|
CREB regulated transcription coactivator 3 |
| chr8_-_64659004 | 0.08 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr18_-_38068456 | 0.08 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr11_-_115310743 | 0.08 |
ENSMUST00000106537.8
ENSMUST00000043931.9 ENSMUST00000073791.10 |
Atp5h
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D |
| chr2_-_122199604 | 0.08 |
ENSMUST00000151130.8
|
Shf
|
Src homology 2 domain containing F |
| chr11_-_69260203 | 0.08 |
ENSMUST00000092971.13
ENSMUST00000108661.8 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr17_-_16046780 | 0.08 |
ENSMUST00000232638.2
ENSMUST00000170578.3 |
Rgmb
|
repulsive guidance molecule family member B |
| chr17_-_65920481 | 0.08 |
ENSMUST00000024897.10
|
Vapa
|
vesicle-associated membrane protein, associated protein A |
| chr5_+_65127412 | 0.08 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr4_+_128777339 | 0.08 |
ENSMUST00000035667.9
|
Trim62
|
tripartite motif-containing 62 |
| chr11_+_68582731 | 0.08 |
ENSMUST00000102611.10
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle |
| chr13_-_58421935 | 0.08 |
ENSMUST00000091579.6
|
Gkap1
|
G kinase anchoring protein 1 |
| chr2_+_156455583 | 0.08 |
ENSMUST00000109567.10
ENSMUST00000169464.9 |
Dlgap4
|
DLG associated protein 4 |
| chr12_+_108601963 | 0.08 |
ENSMUST00000223109.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr6_+_99669640 | 0.08 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr10_+_95350975 | 0.08 |
ENSMUST00000099329.5
|
Ube2n
|
ubiquitin-conjugating enzyme E2N |
| chr5_+_124583524 | 0.08 |
ENSMUST00000100709.7
|
Kmt5a
|
lysine methyltransferase 5A |
| chrX_+_7708295 | 0.08 |
ENSMUST00000115667.10
ENSMUST00000115668.10 ENSMUST00000115665.2 |
Otud5
|
OTU domain containing 5 |
| chr7_-_24937276 | 0.08 |
ENSMUST00000071739.12
ENSMUST00000108411.2 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr7_-_16007542 | 0.07 |
ENSMUST00000169612.3
|
Inafm1
|
InaF motif containing 1 |
| chr3_-_122778052 | 0.07 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr6_+_72074545 | 0.07 |
ENSMUST00000069994.11
ENSMUST00000114112.4 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr14_+_84680993 | 0.07 |
ENSMUST00000071370.7
|
Pcdh17
|
protocadherin 17 |
| chr4_-_22488296 | 0.07 |
ENSMUST00000178174.3
|
Pou3f2
|
POU domain, class 3, transcription factor 2 |
| chr9_+_100956145 | 0.07 |
ENSMUST00000189616.2
|
Msl2
|
MSL complex subunit 2 |
| chr11_-_102298141 | 0.07 |
ENSMUST00000149777.8
ENSMUST00000154001.8 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr8_-_11362731 | 0.07 |
ENSMUST00000033898.10
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr19_+_6291698 | 0.07 |
ENSMUST00000045351.13
|
Atg2a
|
autophagy related 2A |
| chr17_+_25946644 | 0.07 |
ENSMUST00000237183.2
ENSMUST00000237785.2 ENSMUST00000047273.3 |
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr4_+_3938881 | 0.07 |
ENSMUST00000108386.8
ENSMUST00000121110.8 ENSMUST00000149544.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr1_+_152642291 | 0.07 |
ENSMUST00000077755.11
ENSMUST00000097536.6 |
Arpc5
|
actin related protein 2/3 complex, subunit 5 |
| chrX_+_55777139 | 0.07 |
ENSMUST00000023854.10
ENSMUST00000114769.9 |
Fhl1
|
four and a half LIM domains 1 |
| chr16_-_84970580 | 0.07 |
ENSMUST00000227737.2
ENSMUST00000226801.2 |
App
|
amyloid beta (A4) precursor protein |
| chr1_+_60448931 | 0.07 |
ENSMUST00000189082.7
ENSMUST00000187709.7 |
Abi2
|
abl interactor 2 |
| chr1_+_134487928 | 0.07 |
ENSMUST00000112198.3
|
Kdm5b
|
lysine (K)-specific demethylase 5B |
| chr15_-_64184485 | 0.07 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr14_-_30075424 | 0.07 |
ENSMUST00000224198.3
ENSMUST00000238675.2 ENSMUST00000112249.10 ENSMUST00000224785.3 |
Cacna1d
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr4_-_129590372 | 0.07 |
ENSMUST00000137640.3
|
Tmem39b
|
transmembrane protein 39b |
| chr14_+_101891416 | 0.07 |
ENSMUST00000002289.8
|
Uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr9_-_108141105 | 0.07 |
ENSMUST00000166905.8
ENSMUST00000191899.6 |
Dag1
|
dystroglycan 1 |
| chr7_+_44667377 | 0.07 |
ENSMUST00000044111.10
|
Rras
|
related RAS viral (r-ras) oncogene |
| chr10_-_127504416 | 0.07 |
ENSMUST00000129252.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chrX_+_7628891 | 0.07 |
ENSMUST00000077680.10
ENSMUST00000079542.13 ENSMUST00000115679.8 ENSMUST00000137467.8 |
Tfe3
|
transcription factor E3 |
| chr13_-_108026590 | 0.06 |
ENSMUST00000225822.4
ENSMUST00000225197.3 |
Zswim6
|
zinc finger SWIM-type containing 6 |
| chr10_+_19467697 | 0.06 |
ENSMUST00000020188.13
|
Ifngr1
|
interferon gamma receptor 1 |
| chr13_-_69147639 | 0.06 |
ENSMUST00000022013.8
|
Adcy2
|
adenylate cyclase 2 |
| chr11_+_97206542 | 0.06 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr9_-_87613301 | 0.06 |
ENSMUST00000034991.8
|
Tbx18
|
T-box18 |
| chr4_+_53631460 | 0.06 |
ENSMUST00000132151.8
ENSMUST00000159415.9 ENSMUST00000163067.9 |
Fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr2_-_104324745 | 0.06 |
ENSMUST00000028600.14
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr13_+_60749735 | 0.06 |
ENSMUST00000226059.2
ENSMUST00000077453.13 |
Dapk1
|
death associated protein kinase 1 |
| chr9_-_108140925 | 0.06 |
ENSMUST00000171412.7
ENSMUST00000195429.6 ENSMUST00000080435.9 |
Dag1
|
dystroglycan 1 |
| chr10_+_79690452 | 0.06 |
ENSMUST00000165704.8
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr8_+_84699580 | 0.06 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chrX_-_52358663 | 0.06 |
ENSMUST00000114841.2
ENSMUST00000071023.12 |
Fam122b
|
family with sequence similarity 122, member B |
| chr7_+_24920840 | 0.06 |
ENSMUST00000055604.6
|
Zfp526
|
zinc finger protein 526 |
| chr2_+_155850002 | 0.06 |
ENSMUST00000088650.11
|
Ergic3
|
ERGIC and golgi 3 |
| chr17_-_35191127 | 0.06 |
ENSMUST00000087328.4
|
Hspa1a
|
heat shock protein 1A |
| chr15_-_36609208 | 0.06 |
ENSMUST00000001809.15
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr7_+_28508220 | 0.06 |
ENSMUST00000172529.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr13_-_13568106 | 0.06 |
ENSMUST00000021738.10
ENSMUST00000220628.2 |
Gpr137b
|
G protein-coupled receptor 137B |
| chr14_-_79718890 | 0.06 |
ENSMUST00000022601.7
|
Wbp4
|
WW domain binding protein 4 |
| chr13_+_83672389 | 0.06 |
ENSMUST00000200394.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_107438751 | 0.06 |
ENSMUST00000100305.8
ENSMUST00000075012.8 ENSMUST00000106746.8 |
Helz
|
helicase with zinc finger domain |
| chr16_+_4964849 | 0.06 |
ENSMUST00000165810.2
ENSMUST00000230616.2 |
Sec14l5
|
SEC14-like lipid binding 5 |
| chr2_-_122199643 | 0.06 |
ENSMUST00000125826.8
|
Shf
|
Src homology 2 domain containing F |
| chr15_+_79231720 | 0.06 |
ENSMUST00000096350.11
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr7_-_127308059 | 0.06 |
ENSMUST00000061468.9
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr8_+_26275314 | 0.06 |
ENSMUST00000038421.8
|
Lsm1
|
LSM1 homolog, mRNA degradation associated |
| chr16_+_80997580 | 0.06 |
ENSMUST00000037785.14
ENSMUST00000067602.5 |
Ncam2
|
neural cell adhesion molecule 2 |
| chrX_-_160942713 | 0.06 |
ENSMUST00000087085.10
|
Nhs
|
NHS actin remodeling regulator |
| chr9_+_109961048 | 0.06 |
ENSMUST00000088716.12
|
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr18_+_42408418 | 0.06 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr10_-_81186137 | 0.06 |
ENSMUST00000167481.8
|
Hmg20b
|
high mobility group 20B |
| chr12_+_105302853 | 0.06 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr7_-_127307898 | 0.05 |
ENSMUST00000207019.2
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr13_-_12121831 | 0.05 |
ENSMUST00000021750.15
ENSMUST00000170156.3 ENSMUST00000220597.2 |
Ryr2
|
ryanodine receptor 2, cardiac |
| chr17_-_24388384 | 0.05 |
ENSMUST00000024932.12
|
Atp6v0c
|
ATPase, H+ transporting, lysosomal V0 subunit C |
| chr2_+_25070749 | 0.05 |
ENSMUST00000104999.4
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr8_-_25691380 | 0.05 |
ENSMUST00000084512.11
|
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr7_+_80643394 | 0.05 |
ENSMUST00000107353.3
|
Zfp592
|
zinc finger protein 592 |
| chr7_-_44203319 | 0.05 |
ENSMUST00000208366.2
ENSMUST00000207737.2 ENSMUST00000107910.8 ENSMUST00000167197.8 ENSMUST00000128600.3 ENSMUST00000107911.8 ENSMUST00000073488.12 |
Nr1h2
|
nuclear receptor subfamily 1, group H, member 2 |
| chr2_-_181101158 | 0.05 |
ENSMUST00000155535.2
ENSMUST00000029106.13 ENSMUST00000087409.10 |
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr16_+_33504908 | 0.05 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr9_+_109961079 | 0.05 |
ENSMUST00000197480.5
ENSMUST00000197984.5 ENSMUST00000199896.2 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr13_-_55661240 | 0.05 |
ENSMUST00000069929.13
ENSMUST00000069968.13 ENSMUST00000131306.8 ENSMUST00000046246.13 |
Pdlim7
|
PDZ and LIM domain 7 |
| chr11_+_60308077 | 0.05 |
ENSMUST00000070681.7
|
Gid4
|
GID complex subunit 4, VID24 homolog |
| chrX_+_23559282 | 0.05 |
ENSMUST00000035766.13
ENSMUST00000101670.3 |
Wdr44
|
WD repeat domain 44 |
| chr15_+_76131020 | 0.05 |
ENSMUST00000229380.2
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr7_-_127307791 | 0.05 |
ENSMUST00000205977.2
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr14_-_4808744 | 0.05 |
ENSMUST00000022303.17
ENSMUST00000091471.12 |
Thrb
|
thyroid hormone receptor beta |
| chr17_+_56386628 | 0.05 |
ENSMUST00000226053.2
ENSMUST00000002911.10 |
Hdgfl2
|
HDGF like 2 |
| chr9_-_118922534 | 0.05 |
ENSMUST00000010804.4
|
Plcd1
|
phospholipase C, delta 1 |
| chr1_+_171330978 | 0.05 |
ENSMUST00000081527.2
|
Alyref2
|
Aly/REF export factor 2 |
| chr14_-_100521888 | 0.05 |
ENSMUST00000226774.2
|
Klf12
|
Kruppel-like factor 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of cGMP-mediated signaling(GO:0010752) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 0.2 | GO:1904766 | negative regulation of macroautophagy by TORC1 signaling(GO:1904766) |
| 0.1 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 0.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.2 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.4 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.1 | 0.3 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.2 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.2 | GO:0061017 | primary lung bud formation(GO:0060431) hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.0 | 0.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0071874 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 0.1 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0090345 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.6 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) regulation of rRNA processing(GO:2000232) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.1 | GO:0035938 | estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.2 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0070722 | Tle3-Aes complex(GO:0070722) |
| 0.0 | 0.0 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0044547 | rRNA primary transcript binding(GO:0042134) DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.1 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0000401 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |