Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
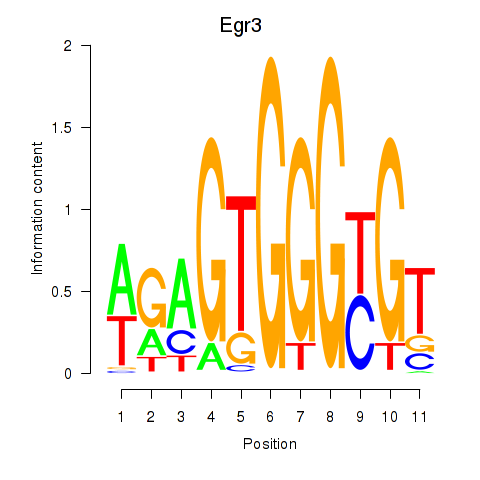
Results for Egr3
Z-value: 0.50
Transcription factors associated with Egr3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr3
|
ENSMUSG00000033730.5 | early growth response 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr3 | mm39_v1_chr14_+_70314652_70314671 | 0.74 | 1.5e-01 | Click! |
Activity profile of Egr3 motif
Sorted Z-values of Egr3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_138278502 | 0.49 |
ENSMUST00000160729.8
|
Stag3
|
stromal antigen 3 |
| chr5_+_138278777 | 0.48 |
ENSMUST00000048028.15
ENSMUST00000162245.8 ENSMUST00000161691.2 |
Stag3
|
stromal antigen 3 |
| chr10_-_68114543 | 0.37 |
ENSMUST00000219238.2
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr17_+_25798059 | 0.31 |
ENSMUST00000141606.3
ENSMUST00000063344.15 ENSMUST00000116641.9 |
Lmf1
|
lipase maturation factor 1 |
| chr1_-_160134873 | 0.30 |
ENSMUST00000193185.6
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr7_+_3648264 | 0.30 |
ENSMUST00000206287.2
ENSMUST00000038913.16 |
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr11_-_69586626 | 0.29 |
ENSMUST00000108649.3
ENSMUST00000174159.8 ENSMUST00000181810.8 |
Tnfsfm13
Tnfsf12
|
tumor necrosis factor (ligand) superfamily, membrane-bound member 13 tumor necrosis factor (ligand) superfamily, member 12 |
| chr2_-_143853122 | 0.26 |
ENSMUST00000016072.12
ENSMUST00000037875.6 |
Rrbp1
|
ribosome binding protein 1 |
| chr2_-_69416365 | 0.22 |
ENSMUST00000100051.9
ENSMUST00000092551.5 ENSMUST00000080953.12 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr12_+_108601963 | 0.20 |
ENSMUST00000223109.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr12_+_49429574 | 0.18 |
ENSMUST00000179669.3
|
Foxg1
|
forkhead box G1 |
| chr5_-_62923463 | 0.16 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr11_-_103158190 | 0.15 |
ENSMUST00000021324.3
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14 |
| chrX_+_100317803 | 0.13 |
ENSMUST00000117203.8
ENSMUST00000087948.11 ENSMUST00000087956.6 |
Med12
|
mediator complex subunit 12 |
| chr9_+_21077010 | 0.13 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr9_-_54956032 | 0.13 |
ENSMUST00000034854.8
|
Chrnb4
|
cholinergic receptor, nicotinic, beta polypeptide 4 |
| chr10_+_3784877 | 0.11 |
ENSMUST00000239159.2
|
Plekhg1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr12_-_52018072 | 0.11 |
ENSMUST00000040583.7
|
Heatr5a
|
HEAT repeat containing 5A |
| chr18_+_65831324 | 0.11 |
ENSMUST00000115097.8
ENSMUST00000117694.2 ENSMUST00000235962.2 |
Oacyl
|
O-acyltransferase like |
| chr12_+_108602008 | 0.11 |
ENSMUST00000172409.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr1_-_74544946 | 0.11 |
ENSMUST00000044260.11
ENSMUST00000186282.7 |
Usp37
|
ubiquitin specific peptidase 37 |
| chr11_-_98666159 | 0.10 |
ENSMUST00000064941.7
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chrX_+_100317629 | 0.09 |
ENSMUST00000117706.8
|
Med12
|
mediator complex subunit 12 |
| chr17_-_46638915 | 0.08 |
ENSMUST00000167360.8
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr1_-_156766957 | 0.08 |
ENSMUST00000171292.8
ENSMUST00000063199.13 ENSMUST00000027886.14 |
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr8_-_94063823 | 0.08 |
ENSMUST00000044602.8
|
Ces1g
|
carboxylesterase 1G |
| chr7_+_48608800 | 0.08 |
ENSMUST00000183659.8
|
Nav2
|
neuron navigator 2 |
| chr17_-_46638874 | 0.07 |
ENSMUST00000047970.14
|
Abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr3_-_126955976 | 0.07 |
ENSMUST00000182994.8
|
Ank2
|
ankyrin 2, brain |
| chr1_-_156766381 | 0.07 |
ENSMUST00000188656.7
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr18_-_38068456 | 0.07 |
ENSMUST00000080033.7
ENSMUST00000115631.8 ENSMUST00000115634.8 |
Diaph1
|
diaphanous related formin 1 |
| chr6_+_103487973 | 0.06 |
ENSMUST00000066905.9
|
Chl1
|
cell adhesion molecule L1-like |
| chr5_+_4073343 | 0.06 |
ENSMUST00000238634.2
|
Akap9
|
A kinase (PRKA) anchor protein (yotiao) 9 |
| chr1_-_164135008 | 0.06 |
ENSMUST00000027866.11
ENSMUST00000120447.8 ENSMUST00000086032.4 |
Blzf1
|
basic leucine zipper nuclear factor 1 |
| chr5_-_138278223 | 0.06 |
ENSMUST00000014089.9
ENSMUST00000161827.8 |
Gpc2
|
glypican 2 (cerebroglycan) |
| chr6_-_88851579 | 0.06 |
ENSMUST00000061262.11
ENSMUST00000140455.8 ENSMUST00000145780.2 |
Podxl2
|
podocalyxin-like 2 |
| chr18_+_53596570 | 0.06 |
ENSMUST00000236096.2
|
Prdm6
|
PR domain containing 6 |
| chr17_+_75772475 | 0.05 |
ENSMUST00000095204.6
|
Rasgrp3
|
RAS, guanyl releasing protein 3 |
| chr16_+_21023505 | 0.05 |
ENSMUST00000006112.7
|
Ephb3
|
Eph receptor B3 |
| chr7_+_109660887 | 0.05 |
ENSMUST00000211798.2
ENSMUST00000084727.11 ENSMUST00000169638.4 |
Zfp143
|
zinc finger protein 143 |
| chr19_+_16933471 | 0.05 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chr18_-_38068430 | 0.05 |
ENSMUST00000025337.14
|
Diaph1
|
diaphanous related formin 1 |
| chr5_-_134776101 | 0.05 |
ENSMUST00000015138.13
|
Eln
|
elastin |
| chr10_-_59057570 | 0.05 |
ENSMUST00000220156.2
ENSMUST00000165971.3 |
Septin10
|
septin 10 |
| chr6_-_124519240 | 0.04 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr12_+_49429790 | 0.04 |
ENSMUST00000021333.5
|
Foxg1
|
forkhead box G1 |
| chr5_-_137101108 | 0.04 |
ENSMUST00000077523.4
ENSMUST00000041388.11 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr7_-_45159790 | 0.03 |
ENSMUST00000211765.2
|
Nucb1
|
nucleobindin 1 |
| chr4_+_40948407 | 0.03 |
ENSMUST00000030128.6
|
Chmp5
|
charged multivesicular body protein 5 |
| chr3_+_106943472 | 0.03 |
ENSMUST00000052718.5
|
Kcna3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr4_+_32238950 | 0.03 |
ENSMUST00000037416.13
|
Bach2
|
BTB and CNC homology, basic leucine zipper transcription factor 2 |
| chr6_-_83549399 | 0.03 |
ENSMUST00000206592.2
ENSMUST00000206400.2 |
Stambp
|
STAM binding protein |
| chr15_+_99122742 | 0.03 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr6_+_113284098 | 0.03 |
ENSMUST00000113122.8
ENSMUST00000204198.3 ENSMUST00000113121.8 ENSMUST00000113119.8 ENSMUST00000113117.4 ENSMUST00000204626.3 ENSMUST00000203577.2 |
Brpf1
|
bromodomain and PHD finger containing, 1 |
| chr17_-_80022480 | 0.02 |
ENSMUST00000234361.2
|
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr12_-_84497718 | 0.02 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr7_+_18810167 | 0.02 |
ENSMUST00000108479.2
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr12_+_86288626 | 0.02 |
ENSMUST00000221368.2
ENSMUST00000071106.6 |
Gpatch2l
|
G patch domain containing 2 like |
| chr4_-_129132963 | 0.02 |
ENSMUST00000097873.10
|
C77080
|
expressed sequence C77080 |
| chr8_+_11606056 | 0.02 |
ENSMUST00000210740.2
ENSMUST00000210670.2 ENSMUST00000054399.6 ENSMUST00000209646.2 |
Ing1
|
inhibitor of growth family, member 1 |
| chr7_-_45159739 | 0.02 |
ENSMUST00000211682.2
ENSMUST00000033096.16 ENSMUST00000209436.2 ENSMUST00000211343.2 |
Nucb1
|
nucleobindin 1 |
| chr10_-_40759307 | 0.02 |
ENSMUST00000044166.9
|
Cdc40
|
cell division cycle 40 |
| chr1_-_156766351 | 0.02 |
ENSMUST00000189648.2
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr5_+_65127412 | 0.02 |
ENSMUST00000031080.15
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr19_-_38113249 | 0.02 |
ENSMUST00000112335.4
|
Rbp4
|
retinol binding protein 4, plasma |
| chr16_+_94171477 | 0.02 |
ENSMUST00000117648.9
ENSMUST00000147352.8 ENSMUST00000150346.8 ENSMUST00000155692.8 ENSMUST00000153988.9 ENSMUST00000139513.9 ENSMUST00000141856.8 ENSMUST00000152117.8 ENSMUST00000150097.8 ENSMUST00000122895.8 ENSMUST00000151770.8 ENSMUST00000231569.2 ENSMUST00000147046.8 ENSMUST00000149885.8 ENSMUST00000127667.8 ENSMUST00000119131.3 ENSMUST00000145883.2 |
Ttc3
|
tetratricopeptide repeat domain 3 |
| chr7_+_144450260 | 0.02 |
ENSMUST00000033389.7
ENSMUST00000207229.2 |
Fgf15
|
fibroblast growth factor 15 |
| chr4_-_40948196 | 0.02 |
ENSMUST00000030125.5
ENSMUST00000108089.8 ENSMUST00000191273.7 |
Bag1
|
BCL2-associated athanogene 1 |
| chr11_+_71641505 | 0.02 |
ENSMUST00000021168.14
|
Wscd1
|
WSC domain containing 1 |
| chr6_+_91661074 | 0.02 |
ENSMUST00000205480.2
ENSMUST00000206545.2 |
Slc6a6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6 |
| chr4_-_129590372 | 0.02 |
ENSMUST00000137640.3
|
Tmem39b
|
transmembrane protein 39b |
| chr4_+_57637817 | 0.01 |
ENSMUST00000150412.4
|
Pakap
|
paralemmin A kinase anchor protein |
| chr5_-_21850579 | 0.01 |
ENSMUST00000051358.11
|
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr11_+_71641806 | 0.01 |
ENSMUST00000108511.8
|
Wscd1
|
WSC domain containing 1 |
| chr19_+_6276408 | 0.01 |
ENSMUST00000025698.14
ENSMUST00000113526.2 |
Gpha2
|
glycoprotein hormone alpha 2 |
| chr7_+_18810097 | 0.01 |
ENSMUST00000032570.14
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif |
| chr16_-_45664591 | 0.01 |
ENSMUST00000076333.12
|
Phldb2
|
pleckstrin homology like domain, family B, member 2 |
| chr9_+_119723931 | 0.01 |
ENSMUST00000036561.8
ENSMUST00000217472.2 ENSMUST00000215307.2 |
Wdr48
|
WD repeat domain 48 |
| chr9_-_66032134 | 0.01 |
ENSMUST00000034946.15
|
Snx1
|
sorting nexin 1 |
| chr11_-_107805830 | 0.01 |
ENSMUST00000039071.3
|
Cacng5
|
calcium channel, voltage-dependent, gamma subunit 5 |
| chr6_+_103488044 | 0.01 |
ENSMUST00000203830.3
|
Chl1
|
cell adhesion molecule L1-like |
| chr7_-_118783820 | 0.01 |
ENSMUST00000084650.6
|
Gpr139
|
G protein-coupled receptor 139 |
| chr1_+_125604706 | 0.01 |
ENSMUST00000027581.7
|
Gpr39
|
G protein-coupled receptor 39 |
| chr6_+_71684846 | 0.01 |
ENSMUST00000212792.2
|
Reep1
|
receptor accessory protein 1 |
| chr9_-_58111589 | 0.01 |
ENSMUST00000217578.2
ENSMUST00000114144.9 ENSMUST00000214649.2 |
Islr
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr1_+_133291302 | 0.01 |
ENSMUST00000135222.9
|
Etnk2
|
ethanolamine kinase 2 |
| chr15_+_81119700 | 0.00 |
ENSMUST00000166855.3
|
Mchr1
|
melanin-concentrating hormone receptor 1 |
| chr10_+_40759468 | 0.00 |
ENSMUST00000019975.14
|
Wasf1
|
WASP family, member 1 |
| chr7_-_142223662 | 0.00 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr7_+_43361930 | 0.00 |
ENSMUST00000066834.8
|
Klk13
|
kallikrein related-peptidase 13 |
| chr10_+_40759815 | 0.00 |
ENSMUST00000105509.2
|
Wasf1
|
WASP family, member 1 |
| chr6_+_103488291 | 0.00 |
ENSMUST00000204321.2
|
Chl1
|
cell adhesion molecule L1-like |
| chr1_+_120268299 | 0.00 |
ENSMUST00000037286.12
|
C1ql2
|
complement component 1, q subcomponent-like 2 |
| chr7_-_103799746 | 0.00 |
ENSMUST00000059121.5
|
Ubqlnl
|
ubiquilin-like |
| chr8_+_63404228 | 0.00 |
ENSMUST00000118003.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr3_+_96736600 | 0.00 |
ENSMUST00000135031.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr4_-_20778530 | 0.00 |
ENSMUST00000119374.8
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr7_-_30754792 | 0.00 |
ENSMUST00000206328.2
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr19_-_38113056 | 0.00 |
ENSMUST00000236283.2
|
Rbp4
|
retinol binding protein 4, plasma |
| chr8_-_49008881 | 0.00 |
ENSMUST00000110345.8
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr3_+_96736774 | 0.00 |
ENSMUST00000138014.8
|
Pdzk1
|
PDZ domain containing 1 |
| chr11_-_68277799 | 0.00 |
ENSMUST00000135141.2
|
Ntn1
|
netrin 1 |
| chr10_+_106306122 | 0.00 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr1_-_161615927 | 0.00 |
ENSMUST00000193648.2
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr11_-_68277631 | 0.00 |
ENSMUST00000021284.4
|
Ntn1
|
netrin 1 |
| chr8_+_63404395 | 0.00 |
ENSMUST00000119068.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr10_+_80134917 | 0.00 |
ENSMUST00000154212.8
|
Apc2
|
APC regulator of WNT signaling pathway 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 | 0.2 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.0 | 0.3 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0090320 | chylomicron assembly(GO:0034378) regulation of chylomicron remnant clearance(GO:0090320) |
| 0.0 | 0.3 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.0 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0000802 | transverse filament(GO:0000802) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |