Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
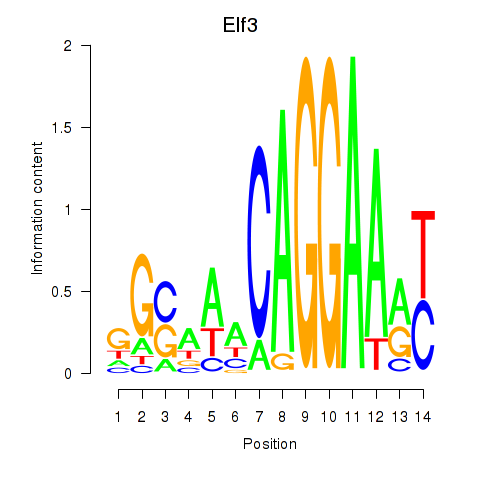
Results for Elf3
Z-value: 1.08
Transcription factors associated with Elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Elf3
|
ENSMUSG00000003051.14 | E74-like factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Elf3 | mm39_v1_chr1_-_135186176_135186306 | 0.89 | 4.5e-02 | Click! |
Activity profile of Elf3 motif
Sorted Z-values of Elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_123195986 | 0.94 |
ENSMUST00000038863.9
ENSMUST00000216843.2 |
Lars2
|
leucyl-tRNA synthetase, mitochondrial |
| chr1_+_16758629 | 0.82 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96 |
| chr2_+_164897547 | 0.75 |
ENSMUST00000017799.12
ENSMUST00000073707.9 |
Cd40
|
CD40 antigen |
| chr13_-_42000958 | 0.61 |
ENSMUST00000072012.10
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr9_-_43027809 | 0.59 |
ENSMUST00000216126.2
ENSMUST00000213544.2 ENSMUST00000061833.6 |
Tlcd5
|
TLC domain containing 5 |
| chr13_-_23727549 | 0.57 |
ENSMUST00000224359.2
|
H2bc9
|
H2B clustered histone 9 |
| chr7_+_101583283 | 0.56 |
ENSMUST00000209639.2
ENSMUST00000210679.2 |
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr2_+_126057020 | 0.50 |
ENSMUST00000164042.3
|
Gm17555
|
predicted gene, 17555 |
| chr1_+_138891155 | 0.49 |
ENSMUST00000200533.5
|
Dennd1b
|
DENN/MADD domain containing 1B |
| chr11_+_32483290 | 0.46 |
ENSMUST00000102821.4
|
Stk10
|
serine/threonine kinase 10 |
| chr3_-_122778052 | 0.45 |
ENSMUST00000199401.2
ENSMUST00000197314.5 ENSMUST00000197934.5 ENSMUST00000090379.7 |
Usp53
|
ubiquitin specific peptidase 53 |
| chr12_+_33365371 | 0.43 |
ENSMUST00000154742.2
|
Atxn7l1
|
ataxin 7-like 1 |
| chr7_+_102246977 | 0.43 |
ENSMUST00000215712.2
|
Olfr552
|
olfactory receptor 552 |
| chr15_-_76079891 | 0.42 |
ENSMUST00000023226.13
|
Plec
|
plectin |
| chr19_+_8828132 | 0.38 |
ENSMUST00000235683.2
ENSMUST00000096257.3 |
Lrrn4cl
|
LRRN4 C-terminal like |
| chr2_-_89951611 | 0.38 |
ENSMUST00000216493.2
ENSMUST00000214404.2 |
Olfr1269
|
olfactory receptor 1269 |
| chr12_-_31549538 | 0.38 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chr13_+_120085355 | 0.38 |
ENSMUST00000099241.4
|
Ccl28
|
chemokine (C-C motif) ligand 28 |
| chr6_-_121450547 | 0.38 |
ENSMUST00000046373.8
ENSMUST00000151397.3 |
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr11_-_62348115 | 0.38 |
ENSMUST00000069456.11
ENSMUST00000018645.13 |
Ncor1
|
nuclear receptor co-repressor 1 |
| chr7_+_110368037 | 0.37 |
ENSMUST00000213373.2
|
Ampd3
|
adenosine monophosphate deaminase 3 |
| chr1_+_34044940 | 0.37 |
ENSMUST00000187486.7
ENSMUST00000182697.8 |
Dst
|
dystonin |
| chr14_+_14159978 | 0.37 |
ENSMUST00000137133.2
ENSMUST00000036070.15 ENSMUST00000121887.8 |
Fam107a
|
family with sequence similarity 107, member A |
| chr10_-_62067026 | 0.36 |
ENSMUST00000047883.11
|
Tspan15
|
tetraspanin 15 |
| chr11_-_48708159 | 0.36 |
ENSMUST00000047145.14
|
Trim41
|
tripartite motif-containing 41 |
| chr11_-_62348599 | 0.35 |
ENSMUST00000127471.9
|
Ncor1
|
nuclear receptor co-repressor 1 |
| chr7_+_101010447 | 0.35 |
ENSMUST00000137384.8
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr8_+_106002772 | 0.34 |
ENSMUST00000014920.8
|
Nol3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr4_+_44756608 | 0.34 |
ENSMUST00000143385.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr19_-_40982576 | 0.34 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr2_+_90229027 | 0.34 |
ENSMUST00000216111.3
|
Olfr1274
|
olfactory receptor 1274 |
| chr18_+_62681982 | 0.34 |
ENSMUST00000055725.12
ENSMUST00000162365.8 |
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr4_+_43730034 | 0.34 |
ENSMUST00000131248.2
|
Spaar
|
small regulatory polypeptide of amino acid response |
| chr3_-_52012462 | 0.33 |
ENSMUST00000121440.4
|
Maml3
|
mastermind like transcriptional coactivator 3 |
| chr9_+_57496725 | 0.33 |
ENSMUST00000053230.7
|
Ulk3
|
unc-51-like kinase 3 |
| chr3_-_15397325 | 0.32 |
ENSMUST00000108361.2
|
Gm9733
|
predicted gene 9733 |
| chr10_+_78816884 | 0.32 |
ENSMUST00000058991.5
ENSMUST00000203973.2 |
Olfr1352
|
olfactory receptor 1352 |
| chr17_+_87061117 | 0.32 |
ENSMUST00000024954.11
|
Epas1
|
endothelial PAS domain protein 1 |
| chr8_-_65582206 | 0.32 |
ENSMUST00000098713.5
|
Smim31
|
small integral membrane protein 31 |
| chr6_-_66759611 | 0.31 |
ENSMUST00000226457.2
|
Vmn1r38
|
vomeronasal 1 receptor 38 |
| chr4_-_139695337 | 0.31 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr9_-_119897358 | 0.30 |
ENSMUST00000064165.5
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr4_-_63321591 | 0.29 |
ENSMUST00000035724.5
|
Akna
|
AT-hook transcription factor |
| chr5_+_115983292 | 0.29 |
ENSMUST00000137952.8
ENSMUST00000148245.8 |
Cit
|
citron |
| chr6_-_129428869 | 0.29 |
ENSMUST00000203162.3
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr2_+_128971620 | 0.28 |
ENSMUST00000035481.5
|
Chchd5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr9_-_119897328 | 0.28 |
ENSMUST00000177637.2
|
Cx3cr1
|
chemokine (C-X3-C motif) receptor 1 |
| chr17_+_47604995 | 0.28 |
ENSMUST00000190020.4
|
Trerf1
|
transcriptional regulating factor 1 |
| chr9_+_117869642 | 0.27 |
ENSMUST00000134433.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr10_-_84276454 | 0.27 |
ENSMUST00000020220.15
|
Nuak1
|
NUAK family, SNF1-like kinase, 1 |
| chr13_-_59970383 | 0.26 |
ENSMUST00000225987.2
|
Tut7
|
terminal uridylyl transferase 7 |
| chr13_-_59970885 | 0.26 |
ENSMUST00000225179.2
ENSMUST00000225576.2 ENSMUST00000071703.6 |
Tut7
|
terminal uridylyl transferase 7 |
| chr9_-_40004854 | 0.26 |
ENSMUST00000050996.6
ENSMUST00000213087.4 |
Olfr983
|
olfactory receptor 983 |
| chr16_-_4698148 | 0.26 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr4_-_156312961 | 0.26 |
ENSMUST00000217885.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr4_+_44756553 | 0.25 |
ENSMUST00000107824.9
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr14_+_32713336 | 0.25 |
ENSMUST00000038956.12
|
Lrrc18
|
leucine rich repeat containing 18 |
| chr19_+_6413703 | 0.25 |
ENSMUST00000131252.8
ENSMUST00000113489.8 |
Sf1
|
splicing factor 1 |
| chr14_+_54598283 | 0.25 |
ENSMUST00000000985.7
|
Oxa1l
|
oxidase assembly 1-like |
| chr7_+_127475968 | 0.25 |
ENSMUST00000131000.2
|
Zfp646
|
zinc finger protein 646 |
| chr7_-_25176959 | 0.24 |
ENSMUST00000098668.3
ENSMUST00000206687.2 ENSMUST00000206676.2 ENSMUST00000205308.2 ENSMUST00000098669.8 ENSMUST00000206171.2 ENSMUST00000098666.9 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr6_-_146403410 | 0.24 |
ENSMUST00000053273.15
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr1_-_156301821 | 0.24 |
ENSMUST00000188027.2
ENSMUST00000187507.7 ENSMUST00000189661.7 |
Soat1
|
sterol O-acyltransferase 1 |
| chr8_+_3403415 | 0.23 |
ENSMUST00000098966.5
ENSMUST00000239102.2 |
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18 |
| chr17_+_21446349 | 0.23 |
ENSMUST00000235895.2
|
Vmn1r234
|
vomeronasal 1 receptor 234 |
| chr9_-_37259664 | 0.23 |
ENSMUST00000216042.2
ENSMUST00000239439.2 ENSMUST00000037275.6 |
Ccdc15
|
coiled-coil domain containing 15 |
| chr7_-_3680530 | 0.23 |
ENSMUST00000038743.15
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr3_-_146205429 | 0.23 |
ENSMUST00000029839.11
|
Spata1
|
spermatogenesis associated 1 |
| chr4_-_156312996 | 0.23 |
ENSMUST00000105571.4
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr5_-_135518098 | 0.22 |
ENSMUST00000201998.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr2_-_86882616 | 0.22 |
ENSMUST00000213781.2
ENSMUST00000217650.2 |
Olfr1106
|
olfactory receptor 1106 |
| chr16_-_59138611 | 0.21 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204 |
| chr9_-_37259690 | 0.21 |
ENSMUST00000213633.2
|
Ccdc15
|
coiled-coil domain containing 15 |
| chr11_+_94900677 | 0.21 |
ENSMUST00000055947.10
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr1_-_121255448 | 0.20 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr15_-_9529898 | 0.20 |
ENSMUST00000228782.2
ENSMUST00000003981.6 |
Il7r
|
interleukin 7 receptor |
| chr1_+_16758731 | 0.20 |
ENSMUST00000190366.2
|
Ly96
|
lymphocyte antigen 96 |
| chr8_+_3543131 | 0.20 |
ENSMUST00000061508.8
ENSMUST00000207318.2 |
Zfp358
|
zinc finger protein 358 |
| chr10_-_61288437 | 0.20 |
ENSMUST00000167087.2
ENSMUST00000020288.15 |
Eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr14_-_70837253 | 0.19 |
ENSMUST00000022690.10
|
Fam160b2
|
family with sequence similarity 160, member B2 |
| chr3_-_113423774 | 0.19 |
ENSMUST00000092154.10
ENSMUST00000106536.8 ENSMUST00000106535.2 |
Rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr9_+_21458138 | 0.19 |
ENSMUST00000034703.15
ENSMUST00000115395.10 ENSMUST00000115394.8 |
Carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr11_+_55104609 | 0.19 |
ENSMUST00000108867.2
|
Slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr13_+_51805745 | 0.19 |
ENSMUST00000110044.2
|
Secisbp2
|
SECIS binding protein 2 |
| chr15_-_80989200 | 0.19 |
ENSMUST00000109579.9
|
Mrtfa
|
myocardin related transcription factor A |
| chr8_+_66838927 | 0.19 |
ENSMUST00000039540.12
ENSMUST00000110253.3 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr6_-_97436223 | 0.19 |
ENSMUST00000113359.8
|
Frmd4b
|
FERM domain containing 4B |
| chr1_+_170104889 | 0.19 |
ENSMUST00000179976.3
|
Sh2d1b1
|
SH2 domain containing 1B1 |
| chr7_+_24476597 | 0.18 |
ENSMUST00000038069.9
ENSMUST00000206847.2 |
Ceacam10
|
carcinoembryonic antigen-related cell adhesion molecule 10 |
| chr13_+_44882998 | 0.18 |
ENSMUST00000174068.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr14_-_20546848 | 0.18 |
ENSMUST00000022353.5
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr7_+_75351713 | 0.18 |
ENSMUST00000207239.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr3_-_105839980 | 0.18 |
ENSMUST00000098758.5
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr4_+_135413593 | 0.18 |
ENSMUST00000074408.7
|
Ifnlr1
|
interferon lambda receptor 1 |
| chr1_-_128520002 | 0.18 |
ENSMUST00000052172.7
ENSMUST00000142893.2 |
Cxcr4
|
chemokine (C-X-C motif) receptor 4 |
| chr7_-_25239229 | 0.18 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr10_-_12424623 | 0.18 |
ENSMUST00000219003.2
|
Utrn
|
utrophin |
| chr2_-_73410632 | 0.17 |
ENSMUST00000028515.4
|
Chrna1
|
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) |
| chr2_+_115412148 | 0.17 |
ENSMUST00000166472.8
ENSMUST00000110918.3 |
Cdin1
|
CDAN1 interacting nuclease 1 |
| chr2_-_151822114 | 0.17 |
ENSMUST00000062047.6
|
Fam110a
|
family with sequence similarity 110, member A |
| chr1_-_80642969 | 0.17 |
ENSMUST00000190983.7
ENSMUST00000191449.2 |
Dock10
|
dedicator of cytokinesis 10 |
| chr3_+_90511068 | 0.17 |
ENSMUST00000001046.7
|
S100a4
|
S100 calcium binding protein A4 |
| chr13_-_74956924 | 0.17 |
ENSMUST00000223206.2
|
Cast
|
calpastatin |
| chr5_+_124577952 | 0.16 |
ENSMUST00000059580.11
|
Kmt5a
|
lysine methyltransferase 5A |
| chr3_+_89325750 | 0.16 |
ENSMUST00000039110.12
ENSMUST00000125036.8 ENSMUST00000191485.7 ENSMUST00000154791.8 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr10_+_76284907 | 0.16 |
ENSMUST00000092406.12
|
2610028H24Rik
|
RIKEN cDNA 2610028H24 gene |
| chr5_-_109704339 | 0.16 |
ENSMUST00000198960.2
|
Crlf2
|
cytokine receptor-like factor 2 |
| chr11_+_67345895 | 0.16 |
ENSMUST00000108681.9
|
Gas7
|
growth arrest specific 7 |
| chr7_-_12829100 | 0.16 |
ENSMUST00000209822.3
ENSMUST00000235753.2 |
Vmn1r85
|
vomeronasal 1 receptor 85 |
| chr7_+_103893658 | 0.16 |
ENSMUST00000106849.9
ENSMUST00000060315.12 |
Trim34a
|
tripartite motif-containing 34A |
| chr4_-_118266416 | 0.16 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr5_-_135494775 | 0.16 |
ENSMUST00000212301.2
|
Hip1
|
huntingtin interacting protein 1 |
| chr4_-_116413092 | 0.16 |
ENSMUST00000069674.6
ENSMUST00000106478.9 |
Tmem69
|
transmembrane protein 69 |
| chr1_+_40363701 | 0.16 |
ENSMUST00000095020.9
ENSMUST00000194296.6 |
Il1rl2
|
interleukin 1 receptor-like 2 |
| chr9_+_20555629 | 0.15 |
ENSMUST00000161887.8
|
Ubl5
|
ubiquitin-like 5 |
| chr18_-_35348049 | 0.15 |
ENSMUST00000091636.5
ENSMUST00000236680.2 |
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr6_-_116050081 | 0.15 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr19_+_26600820 | 0.15 |
ENSMUST00000176584.2
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_+_12212220 | 0.15 |
ENSMUST00000098822.10
ENSMUST00000123589.8 |
Zfp606
|
zinc finger protein 606 |
| chr8_+_123980934 | 0.14 |
ENSMUST00000001092.15
|
Zfp276
|
zinc finger protein (C2H2 type) 276 |
| chr13_+_110039620 | 0.14 |
ENSMUST00000120664.8
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr12_-_112824506 | 0.14 |
ENSMUST00000021729.9
|
Gpr132
|
G protein-coupled receptor 132 |
| chr7_-_101582987 | 0.14 |
ENSMUST00000106964.8
ENSMUST00000106963.2 ENSMUST00000078448.11 ENSMUST00000106966.8 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr19_-_17333972 | 0.13 |
ENSMUST00000174236.8
|
Gcnt1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr6_+_83214357 | 0.13 |
ENSMUST00000039212.8
ENSMUST00000113899.8 |
Slc4a5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5 |
| chr11_+_75570085 | 0.13 |
ENSMUST00000017920.14
ENSMUST00000108426.8 ENSMUST00000108425.8 ENSMUST00000093115.4 |
Crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr7_-_80051455 | 0.13 |
ENSMUST00000120753.3
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr9_-_37259629 | 0.13 |
ENSMUST00000217238.3
|
Ccdc15
|
coiled-coil domain containing 15 |
| chr1_-_121255400 | 0.13 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr11_-_97886997 | 0.12 |
ENSMUST00000042971.16
|
Arl5c
|
ADP-ribosylation factor-like 5C |
| chr16_-_55755160 | 0.12 |
ENSMUST00000122280.8
ENSMUST00000121703.3 |
Cep97
|
centrosomal protein 97 |
| chr1_-_80191649 | 0.12 |
ENSMUST00000058748.2
|
Fam124b
|
family with sequence similarity 124, member B |
| chr17_+_31276649 | 0.12 |
ENSMUST00000236391.2
ENSMUST00000024829.8 ENSMUST00000236427.2 |
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr7_+_103893672 | 0.12 |
ENSMUST00000106848.8
|
Trim34a
|
tripartite motif-containing 34A |
| chr7_-_127475949 | 0.12 |
ENSMUST00000106262.2
ENSMUST00000106263.8 ENSMUST00000054415.12 |
Zfp668
|
zinc finger protein 668 |
| chr15_-_64184485 | 0.12 |
ENSMUST00000177083.8
ENSMUST00000177371.8 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr16_-_3535958 | 0.12 |
ENSMUST00000023180.15
|
Mefv
|
Mediterranean fever |
| chr6_-_146403638 | 0.12 |
ENSMUST00000079573.13
|
Itpr2
|
inositol 1,4,5-triphosphate receptor 2 |
| chr1_-_133834790 | 0.12 |
ENSMUST00000149380.8
ENSMUST00000124051.9 |
Optc
|
opticin |
| chr11_+_115054157 | 0.11 |
ENSMUST00000021077.4
|
Slc9a3r1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
| chr6_+_70648743 | 0.11 |
ENSMUST00000103401.3
|
Igkv3-4
|
immunoglobulin kappa variable 3-4 |
| chr11_+_51475584 | 0.11 |
ENSMUST00000167797.8
ENSMUST00000020625.7 |
Phykpl
|
5-phosphohydroxy-L-lysine phospholyase |
| chr3_+_84573499 | 0.11 |
ENSMUST00000107682.2
|
Tmem154
|
transmembrane protein 154 |
| chr10_+_127928622 | 0.11 |
ENSMUST00000219072.2
ENSMUST00000045621.9 ENSMUST00000170054.9 |
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr2_-_45001141 | 0.10 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_192855776 | 0.10 |
ENSMUST00000161235.3
ENSMUST00000160077.2 ENSMUST00000178744.2 ENSMUST00000192189.2 ENSMUST00000110831.4 ENSMUST00000191613.2 |
A130010J15Rik
|
RIKEN cDNA A130010J15 gene |
| chr8_+_110505494 | 0.10 |
ENSMUST00000034171.9
|
Ap1g1
|
adaptor protein complex AP-1, gamma 1 subunit |
| chr3_+_89325901 | 0.10 |
ENSMUST00000128238.8
ENSMUST00000107417.9 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr1_-_160958998 | 0.10 |
ENSMUST00000111611.8
|
Klhl20
|
kelch-like 20 |
| chr6_+_54794433 | 0.10 |
ENSMUST00000127331.2
|
Znrf2
|
zinc and ring finger 2 |
| chr16_-_3535940 | 0.10 |
ENSMUST00000229725.2
ENSMUST00000100222.4 |
Mefv
|
Mediterranean fever |
| chr11_-_103235475 | 0.10 |
ENSMUST00000041385.14
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr14_+_20757615 | 0.10 |
ENSMUST00000022358.9
ENSMUST00000224751.2 |
Zswim8
|
zinc finger SWIM-type containing 8 |
| chr1_+_186947683 | 0.10 |
ENSMUST00000065573.14
ENSMUST00000110943.9 ENSMUST00000044812.12 |
Gpatch2
|
G patch domain containing 2 |
| chr4_-_122779837 | 0.10 |
ENSMUST00000106255.8
ENSMUST00000106257.10 |
Cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr3_+_87283687 | 0.10 |
ENSMUST00000163661.8
ENSMUST00000072480.9 |
Fcrl1
|
Fc receptor-like 1 |
| chr19_-_12901783 | 0.09 |
ENSMUST00000213713.2
ENSMUST00000216888.2 ENSMUST00000213177.2 |
Olfr1448
|
olfactory receptor 1448 |
| chr17_-_35392254 | 0.09 |
ENSMUST00000025257.12
|
Aif1
|
allograft inflammatory factor 1 |
| chr12_+_36431449 | 0.09 |
ENSMUST00000221452.2
ENSMUST00000062041.6 ENSMUST00000220519.2 ENSMUST00000221895.2 |
Crppa
|
CDP-L-ribitol pyrophosphorylase A |
| chr1_+_72750418 | 0.09 |
ENSMUST00000059980.11
|
Rpl37a
|
ribosomal protein L37a |
| chr5_-_142594549 | 0.09 |
ENSMUST00000037048.9
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr2_-_84545504 | 0.09 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr8_+_93687561 | 0.09 |
ENSMUST00000072939.8
|
Slc6a2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| chr8_+_73373356 | 0.09 |
ENSMUST00000161254.9
|
Nwd1
|
NACHT and WD repeat domain containing 1 |
| chr9_-_116004265 | 0.09 |
ENSMUST00000061101.12
|
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr3_+_146205562 | 0.09 |
ENSMUST00000090031.12
ENSMUST00000118280.2 |
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr10_+_79762858 | 0.09 |
ENSMUST00000019708.12
ENSMUST00000105377.8 |
Arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr1_+_170136372 | 0.09 |
ENSMUST00000056991.6
|
Spata46
|
spermatogenesis associated 46 |
| chr17_-_79142753 | 0.09 |
ENSMUST00000097281.4
|
Heatr5b
|
HEAT repeat containing 5B |
| chr9_+_117869543 | 0.09 |
ENSMUST00000044454.12
|
Azi2
|
5-azacytidine induced gene 2 |
| chr9_-_56835633 | 0.09 |
ENSMUST00000050916.7
|
Snx33
|
sorting nexin 33 |
| chr18_-_38471962 | 0.08 |
ENSMUST00000139885.2
ENSMUST00000235590.2 ENSMUST00000237487.2 ENSMUST00000063814.15 |
Gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr1_+_58834532 | 0.08 |
ENSMUST00000027189.15
|
Casp8
|
caspase 8 |
| chr11_-_75345482 | 0.08 |
ENSMUST00000173320.8
|
Wdr81
|
WD repeat domain 81 |
| chrX_-_7956682 | 0.08 |
ENSMUST00000033505.7
|
Was
|
Wiskott-Aldrich syndrome |
| chr1_+_58841650 | 0.08 |
ENSMUST00000165549.8
|
Casp8
|
caspase 8 |
| chr7_-_127897253 | 0.08 |
ENSMUST00000033044.16
|
Rusf1
|
RUS family member 1 |
| chr9_+_117869577 | 0.08 |
ENSMUST00000133580.8
|
Azi2
|
5-azacytidine induced gene 2 |
| chr8_-_71848429 | 0.08 |
ENSMUST00000049184.9
|
Ushbp1
|
USH1 protein network component harmonin binding protein 1 |
| chr15_-_78687216 | 0.08 |
ENSMUST00000164826.8
|
Card10
|
caspase recruitment domain family, member 10 |
| chr16_+_18317613 | 0.08 |
ENSMUST00000149035.8
ENSMUST00000167778.9 ENSMUST00000090086.11 ENSMUST00000115601.8 ENSMUST00000147739.8 ENSMUST00000146673.2 |
Gnb1l
Rtl10
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like retrotransposon Gag like 10 |
| chr3_-_88362606 | 0.08 |
ENSMUST00000125526.8
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr14_+_32713349 | 0.07 |
ENSMUST00000120866.8
ENSMUST00000120588.8 |
Lrrc18
|
leucine rich repeat containing 18 |
| chr9_+_44990447 | 0.07 |
ENSMUST00000050020.8
|
Jaml
|
junction adhesion molecule like |
| chr9_-_72399221 | 0.07 |
ENSMUST00000185151.8
ENSMUST00000085358.12 ENSMUST00000184125.8 ENSMUST00000183574.8 ENSMUST00000184831.8 |
Tex9
|
testis expressed gene 9 |
| chr5_+_145063568 | 0.07 |
ENSMUST00000138922.2
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chrX_-_43457202 | 0.07 |
ENSMUST00000115057.2
|
Dcaf12l2
|
DDB1 and CUL4 associated factor 12-like 2 |
| chr1_-_80643024 | 0.07 |
ENSMUST00000187774.7
|
Dock10
|
dedicator of cytokinesis 10 |
| chr6_+_29694181 | 0.07 |
ENSMUST00000046750.14
ENSMUST00000115250.4 |
Tspan33
|
tetraspanin 33 |
| chr11_+_46345784 | 0.07 |
ENSMUST00000109229.2
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr5_-_136227760 | 0.07 |
ENSMUST00000149151.2
ENSMUST00000151786.8 |
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chr5_-_137869969 | 0.07 |
ENSMUST00000196162.5
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr5_-_3691453 | 0.07 |
ENSMUST00000140871.2
|
Gatad1
|
GATA zinc finger domain containing 1 |
| chr7_+_101010672 | 0.07 |
ENSMUST00000138628.8
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_-_192883642 | 0.06 |
ENSMUST00000192020.6
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr14_+_20724378 | 0.06 |
ENSMUST00000224492.2
ENSMUST00000223751.2 ENSMUST00000225108.2 ENSMUST00000224754.2 |
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr12_-_100486950 | 0.06 |
ENSMUST00000223020.2
ENSMUST00000062957.8 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr5_+_64316771 | 0.06 |
ENSMUST00000043893.13
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr1_-_133617824 | 0.06 |
ENSMUST00000189524.2
ENSMUST00000169295.8 |
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr5_-_143166476 | 0.06 |
ENSMUST00000049861.11
ENSMUST00000165318.4 |
Rbak
|
RB-associated KRAB zinc finger |
Network of associatons between targets according to the STRING database.
First level regulatory network of Elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0002880 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.2 | 0.7 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.2 | 0.5 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 1.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.6 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.1 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.4 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.3 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.2 | GO:0071640 | regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.1 | 0.4 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.3 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.7 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.2 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.0 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.3 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.5 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.0 | 0.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.0 | 0.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.0 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0002856 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.2 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.1 | GO:0055099 | glycoprotein transport(GO:0034436) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.0 | GO:0032487 | cerebellar cortex structural organization(GO:0021698) regulation of Rap protein signal transduction(GO:0032487) negative regulation of integrin activation(GO:0033624) |
| 0.0 | 0.0 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.0 | 0.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.0 | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:1900190 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.6 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.2 | 1.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.2 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.1 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0061749 | forked DNA-dependent helicase activity(GO:0061749) flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |