Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Emx1_Emx2
Z-value: 1.70
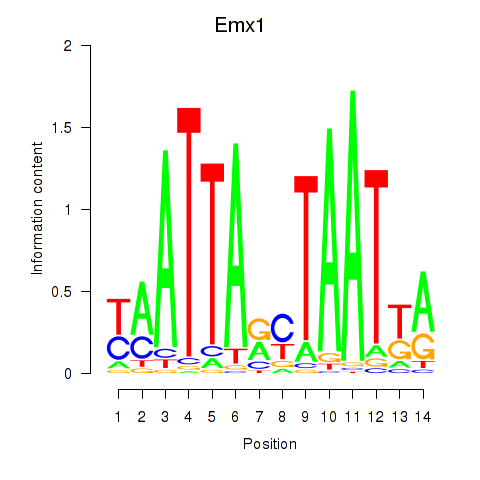
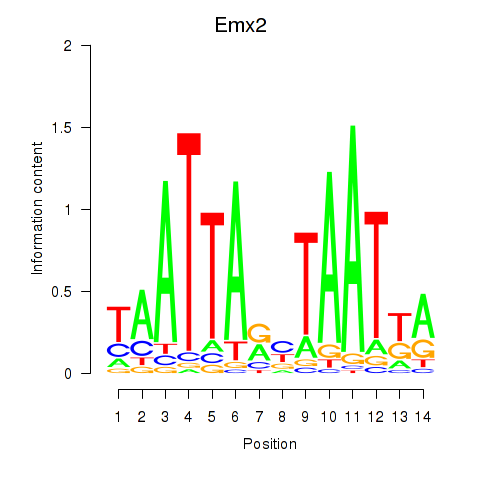
Transcription factors associated with Emx1_Emx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Emx1
|
ENSMUSG00000033726.9 | empty spiracles homeobox 1 |
|
Emx2
|
ENSMUSG00000043969.5 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Emx1 | mm39_v1_chr6_+_85164420_85164505 | -0.66 | 2.3e-01 | Click! |
| Emx2 | mm39_v1_chr19_+_59446804_59446883 | -0.37 | 5.4e-01 | Click! |
Activity profile of Emx1_Emx2 motif
Sorted Z-values of Emx1_Emx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_86587728 | 1.85 |
ENSMUST00000149700.8
|
Plin2
|
perilipin 2 |
| chr10_+_81093110 | 1.65 |
ENSMUST00000117488.8
ENSMUST00000105328.10 ENSMUST00000121205.8 |
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr14_+_69585036 | 1.54 |
ENSMUST00000064831.6
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr12_-_75678092 | 1.46 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr17_-_36343573 | 1.32 |
ENSMUST00000102678.5
|
H2-T23
|
histocompatibility 2, T region locus 23 |
| chr2_+_36120438 | 1.31 |
ENSMUST00000062069.6
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr2_-_157408239 | 1.15 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr18_-_43610829 | 1.14 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr9_+_21634779 | 1.09 |
ENSMUST00000034713.9
|
Ldlr
|
low density lipoprotein receptor |
| chr3_-_106126794 | 1.09 |
ENSMUST00000082219.6
|
Chil4
|
chitinase-like 4 |
| chr2_-_86180622 | 1.02 |
ENSMUST00000099894.5
ENSMUST00000213564.3 |
Olfr1055
|
olfactory receptor 1055 |
| chr13_+_38223023 | 0.87 |
ENSMUST00000226110.2
|
Riok1
|
RIO kinase 1 |
| chr2_-_93283024 | 0.80 |
ENSMUST00000111257.8
ENSMUST00000145553.8 |
Cd82
|
CD82 antigen |
| chr15_+_65682066 | 0.79 |
ENSMUST00000211878.2
|
Efr3a
|
EFR3 homolog A |
| chr1_-_133537953 | 0.75 |
ENSMUST00000164574.2
ENSMUST00000166291.8 ENSMUST00000164096.2 ENSMUST00000166915.8 |
Snrpe
|
small nuclear ribonucleoprotein E |
| chr19_-_24178000 | 0.74 |
ENSMUST00000233658.3
|
Tjp2
|
tight junction protein 2 |
| chr15_+_100202061 | 0.66 |
ENSMUST00000229574.2
ENSMUST00000229217.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr14_-_104760051 | 0.63 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr15_+_100202021 | 0.61 |
ENSMUST00000230472.2
|
Mettl7a1
|
methyltransferase like 7A1 |
| chr4_+_150321142 | 0.60 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr15_+_100202079 | 0.60 |
ENSMUST00000230252.2
ENSMUST00000231166.2 |
Mettl7a1
|
methyltransferase like 7A1 |
| chr11_+_109434519 | 0.60 |
ENSMUST00000106696.2
|
Arsg
|
arylsulfatase G |
| chr2_-_151586063 | 0.60 |
ENSMUST00000109869.2
|
Psmf1
|
proteasome (prosome, macropain) inhibitor subunit 1 |
| chr11_+_17207558 | 0.58 |
ENSMUST00000000594.9
ENSMUST00000156784.2 |
C1d
|
C1D nuclear receptor co-repressor |
| chr9_+_5298669 | 0.58 |
ENSMUST00000238505.2
|
Casp1
|
caspase 1 |
| chr4_-_150998857 | 0.56 |
ENSMUST00000105675.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr12_-_55045887 | 0.56 |
ENSMUST00000173529.2
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A |
| chr8_-_4829519 | 0.55 |
ENSMUST00000022945.9
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr2_+_128942900 | 0.55 |
ENSMUST00000103205.11
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr4_-_133972890 | 0.54 |
ENSMUST00000030644.8
|
Zfp593
|
zinc finger protein 593 |
| chr17_-_74602469 | 0.54 |
ENSMUST00000233144.2
|
Memo1
|
mediator of cell motility 1 |
| chr12_-_84664001 | 0.53 |
ENSMUST00000221070.2
ENSMUST00000021666.6 ENSMUST00000223107.2 |
Abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr2_+_128942919 | 0.53 |
ENSMUST00000028874.8
|
Polr1b
|
polymerase (RNA) I polypeptide B |
| chr3_+_106020545 | 0.51 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chr11_-_79853200 | 0.50 |
ENSMUST00000108241.8
ENSMUST00000043152.6 |
Utp6
|
UTP6 small subunit processome component |
| chr2_+_30331839 | 0.50 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr10_-_126866682 | 0.50 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr8_-_96615138 | 0.48 |
ENSMUST00000034097.8
|
Got2
|
glutamatic-oxaloacetic transaminase 2, mitochondrial |
| chr1_-_63215812 | 0.46 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr8_-_4829473 | 0.44 |
ENSMUST00000207262.2
|
Shcbp1
|
Shc SH2-domain binding protein 1 |
| chr14_+_118374511 | 0.43 |
ENSMUST00000022728.4
|
Gpr180
|
G protein-coupled receptor 180 |
| chr6_+_17749169 | 0.43 |
ENSMUST00000053148.14
ENSMUST00000115417.4 |
St7
|
suppression of tumorigenicity 7 |
| chr9_+_69919822 | 0.42 |
ENSMUST00000118198.8
ENSMUST00000119905.8 ENSMUST00000119413.8 ENSMUST00000140305.8 ENSMUST00000122087.8 |
Gtf2a2
|
general transcription factor II A, 2 |
| chr15_-_36496880 | 0.42 |
ENSMUST00000228601.2
ENSMUST00000057486.9 |
Ankrd46
|
ankyrin repeat domain 46 |
| chr11_-_106889291 | 0.40 |
ENSMUST00000124541.8
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chrX_-_8059597 | 0.39 |
ENSMUST00000143223.2
ENSMUST00000033509.15 |
Ebp
|
phenylalkylamine Ca2+ antagonist (emopamil) binding protein |
| chr7_-_121700958 | 0.39 |
ENSMUST00000139456.2
ENSMUST00000106471.9 ENSMUST00000123296.8 ENSMUST00000033157.10 |
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr3_-_130524024 | 0.38 |
ENSMUST00000079085.11
|
Rpl34
|
ribosomal protein L34 |
| chr6_+_134617903 | 0.37 |
ENSMUST00000062755.10
|
Borcs5
|
BLOC-1 related complex subunit 5 |
| chr7_-_78432774 | 0.37 |
ENSMUST00000032841.7
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr4_-_92079986 | 0.37 |
ENSMUST00000123179.2
|
Gm12666
|
predicted gene 12666 |
| chrX_-_158921370 | 0.37 |
ENSMUST00000033662.9
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1 |
| chr6_+_66873381 | 0.36 |
ENSMUST00000043148.13
ENSMUST00000114228.8 ENSMUST00000114227.8 ENSMUST00000114226.8 ENSMUST00000204511.3 ENSMUST00000114225.8 ENSMUST00000114224.8 ENSMUST00000114222.4 |
Gng12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr18_+_31742565 | 0.36 |
ENSMUST00000164667.2
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr11_+_6510167 | 0.36 |
ENSMUST00000109722.9
|
Ccm2
|
cerebral cavernous malformation 2 |
| chr7_+_131162338 | 0.35 |
ENSMUST00000208571.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr6_-_89572629 | 0.35 |
ENSMUST00000113550.6
ENSMUST00000032172.14 |
Chchd6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr17_+_64203017 | 0.34 |
ENSMUST00000000129.14
|
Fer
|
fer (fms/fps related) protein kinase |
| chr16_-_65359406 | 0.34 |
ENSMUST00000231259.2
|
Chmp2b
|
charged multivesicular body protein 2B |
| chr5_+_147456497 | 0.33 |
ENSMUST00000175807.8
|
Pan3
|
PAN3 poly(A) specific ribonuclease subunit |
| chr13_-_4659120 | 0.33 |
ENSMUST00000091848.7
ENSMUST00000110691.10 |
Akr1e1
|
aldo-keto reductase family 1, member E1 |
| chr4_-_107975723 | 0.32 |
ENSMUST00000030340.15
|
Scp2
|
sterol carrier protein 2, liver |
| chr12_+_87490666 | 0.32 |
ENSMUST00000161023.8
ENSMUST00000160488.8 ENSMUST00000077462.8 ENSMUST00000160880.2 |
Slirp
|
SRA stem-loop interacting RNA binding protein |
| chr2_-_87504008 | 0.32 |
ENSMUST00000213835.2
|
Olfr1135
|
olfactory receptor 1135 |
| chr2_+_151385797 | 0.32 |
ENSMUST00000142271.2
|
Fkbp1a
|
FK506 binding protein 1a |
| chr5_+_45650716 | 0.32 |
ENSMUST00000046122.11
|
Lap3
|
leucine aminopeptidase 3 |
| chr2_-_92876398 | 0.32 |
ENSMUST00000111272.3
ENSMUST00000178666.8 ENSMUST00000147339.3 |
Prdm11
|
PR domain containing 11 |
| chr17_+_29493113 | 0.31 |
ENSMUST00000234326.2
ENSMUST00000235117.2 |
BC004004
|
cDNA sequence BC004004 |
| chr1_+_179788037 | 0.31 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr11_+_96209093 | 0.30 |
ENSMUST00000049241.9
|
Hoxb4
|
homeobox B4 |
| chr12_+_84332006 | 0.30 |
ENSMUST00000123614.8
ENSMUST00000147363.8 ENSMUST00000135001.8 ENSMUST00000146377.8 |
Ptgr2
|
prostaglandin reductase 2 |
| chr19_+_34194990 | 0.29 |
ENSMUST00000119603.2
|
Stambpl1
|
STAM binding protein like 1 |
| chr7_+_43321426 | 0.29 |
ENSMUST00000038332.9
|
Ctu1
|
cytosolic thiouridylase subunit 1 |
| chr14_+_48683797 | 0.29 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chr6_+_142702403 | 0.28 |
ENSMUST00000032419.9
|
Cmas
|
cytidine monophospho-N-acetylneuraminic acid synthetase |
| chr7_+_131162137 | 0.27 |
ENSMUST00000207231.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr6_-_13607963 | 0.27 |
ENSMUST00000031554.9
ENSMUST00000149123.3 |
Tmem168
|
transmembrane protein 168 |
| chr1_-_63215952 | 0.27 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chrX_+_158086253 | 0.27 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr17_+_29493049 | 0.27 |
ENSMUST00000149405.4
|
BC004004
|
cDNA sequence BC004004 |
| chr17_-_56343531 | 0.27 |
ENSMUST00000233803.2
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr13_-_43634695 | 0.27 |
ENSMUST00000144326.4
|
Ranbp9
|
RAN binding protein 9 |
| chr4_-_82768958 | 0.26 |
ENSMUST00000139401.2
|
Zdhhc21
|
zinc finger, DHHC domain containing 21 |
| chr4_+_126156118 | 0.26 |
ENSMUST00000030660.9
|
Trappc3
|
trafficking protein particle complex 3 |
| chr10_-_80754016 | 0.26 |
ENSMUST00000057623.14
ENSMUST00000179022.8 |
Lmnb2
|
lamin B2 |
| chr7_+_126575781 | 0.26 |
ENSMUST00000206450.2
ENSMUST00000205830.2 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr5_-_65248962 | 0.26 |
ENSMUST00000212080.2
|
Tmem156
|
transmembrane protein 156 |
| chr10_+_128158413 | 0.26 |
ENSMUST00000219836.2
|
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr5_-_139330906 | 0.26 |
ENSMUST00000049630.13
|
Cox19
|
cytochrome c oxidase assembly protein 19 |
| chr7_+_78432867 | 0.26 |
ENSMUST00000032840.5
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr2_+_85809620 | 0.26 |
ENSMUST00000056849.3
|
Olfr1030
|
olfactory receptor 1030 |
| chr19_-_33728759 | 0.25 |
ENSMUST00000147153.4
|
Lipo2
|
lipase, member O2 |
| chr2_-_17465410 | 0.25 |
ENSMUST00000145492.2
|
Nebl
|
nebulette |
| chr14_+_55797468 | 0.25 |
ENSMUST00000147981.2
ENSMUST00000133256.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr12_+_71021395 | 0.24 |
ENSMUST00000160027.8
ENSMUST00000160864.8 |
Psma3
|
proteasome subunit alpha 3 |
| chr17_+_56312672 | 0.24 |
ENSMUST00000133998.8
|
Mpnd
|
MPN domain containing |
| chr2_-_155771938 | 0.23 |
ENSMUST00000152766.8
ENSMUST00000139232.8 ENSMUST00000109632.8 ENSMUST00000006036.13 ENSMUST00000142655.2 ENSMUST00000159238.2 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr3_+_32490300 | 0.22 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr12_+_80509869 | 0.22 |
ENSMUST00000038185.10
|
Exd2
|
exonuclease 3'-5' domain containing 2 |
| chr2_-_119493237 | 0.22 |
ENSMUST00000028768.2
ENSMUST00000110801.8 ENSMUST00000110802.8 |
Ndufaf1
|
NADH:ubiquinone oxidoreductase complex assembly factor 1 |
| chr15_+_99192968 | 0.22 |
ENSMUST00000128352.8
ENSMUST00000145482.8 |
Prpf40b
|
pre-mRNA processing factor 40B |
| chr12_-_56392646 | 0.22 |
ENSMUST00000021416.9
|
Mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr11_+_31823096 | 0.22 |
ENSMUST00000155278.2
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr15_+_6673167 | 0.21 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein |
| chr2_+_144435974 | 0.21 |
ENSMUST00000136628.2
|
Smim26
|
small integral membrane protein 26 |
| chr9_-_15191218 | 0.21 |
ENSMUST00000034411.10
|
Med17
|
mediator complex subunit 17 |
| chr17_-_24698007 | 0.21 |
ENSMUST00000234335.2
ENSMUST00000234686.2 ENSMUST00000234941.2 ENSMUST00000234543.2 ENSMUST00000179163.3 ENSMUST00000070888.14 |
Mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr4_+_102446883 | 0.21 |
ENSMUST00000097949.11
ENSMUST00000106901.2 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr1_+_75498162 | 0.21 |
ENSMUST00000027414.16
ENSMUST00000113553.2 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr2_-_155772110 | 0.21 |
ENSMUST00000109636.11
ENSMUST00000109631.8 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr4_-_149184259 | 0.20 |
ENSMUST00000103217.11
|
Pex14
|
peroxisomal biogenesis factor 14 |
| chr7_-_51511964 | 0.20 |
ENSMUST00000169357.2
|
Fancf
|
Fanconi anemia, complementation group F |
| chr2_-_34716083 | 0.20 |
ENSMUST00000113077.8
ENSMUST00000028220.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr2_+_110551685 | 0.20 |
ENSMUST00000111016.9
|
Muc15
|
mucin 15 |
| chr10_-_126866658 | 0.19 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr5_+_44070486 | 0.19 |
ENSMUST00000122204.3
ENSMUST00000200338.2 |
Gm7879
|
predicted pseudogene 7879 |
| chr1_+_179788675 | 0.19 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr18_-_66135683 | 0.19 |
ENSMUST00000120461.9
ENSMUST00000048260.15 ENSMUST00000236866.2 |
Lman1
|
lectin, mannose-binding, 1 |
| chr17_+_29493157 | 0.19 |
ENSMUST00000234234.2
|
BC004004
|
cDNA sequence BC004004 |
| chr5_-_65248927 | 0.19 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr6_+_67838100 | 0.19 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr9_+_78099229 | 0.19 |
ENSMUST00000034903.7
|
Gsta4
|
glutathione S-transferase, alpha 4 |
| chr6_+_129510117 | 0.19 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr7_+_126575752 | 0.18 |
ENSMUST00000206346.2
|
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chrX_-_55643429 | 0.18 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr12_-_87490580 | 0.17 |
ENSMUST00000162961.8
ENSMUST00000185301.2 |
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr4_+_154059619 | 0.17 |
ENSMUST00000047497.15
|
Cep104
|
centrosomal protein 104 |
| chr1_-_84817000 | 0.17 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr17_-_45910529 | 0.16 |
ENSMUST00000171847.8
ENSMUST00000166633.8 ENSMUST00000169729.8 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr8_-_108315024 | 0.16 |
ENSMUST00000044106.6
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr7_+_131162439 | 0.16 |
ENSMUST00000207442.2
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr15_-_76491758 | 0.16 |
ENSMUST00000071898.7
|
Cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chrX_+_151922936 | 0.16 |
ENSMUST00000039720.11
ENSMUST00000144175.3 |
Rragb
|
Ras-related GTP binding B |
| chr6_-_115014777 | 0.16 |
ENSMUST00000174848.8
ENSMUST00000032461.12 |
Tamm41
|
TAM41 mitochondrial translocator assembly and maintenance homolog |
| chr7_+_89814713 | 0.16 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_-_33764859 | 0.16 |
ENSMUST00000148137.9
|
Lipo1
|
lipase, member O1 |
| chr17_+_37274714 | 0.16 |
ENSMUST00000209623.2
|
Polr1has
|
RNA polymerase I subunit H, antisense |
| chr2_+_121337226 | 0.16 |
ENSMUST00000099473.10
ENSMUST00000110602.9 |
Wdr76
|
WD repeat domain 76 |
| chr4_-_11254253 | 0.16 |
ENSMUST00000044616.10
ENSMUST00000108319.9 ENSMUST00000108318.3 |
Ints8
|
integrator complex subunit 8 |
| chr9_+_111014254 | 0.16 |
ENSMUST00000198986.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_+_110551927 | 0.15 |
ENSMUST00000111017.9
|
Muc15
|
mucin 15 |
| chr6_-_57512355 | 0.15 |
ENSMUST00000042766.6
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr8_-_8740471 | 0.15 |
ENSMUST00000048545.10
|
Arglu1
|
arginine and glutamate rich 1 |
| chr3_-_37180093 | 0.15 |
ENSMUST00000029275.6
|
Il2
|
interleukin 2 |
| chr1_-_37535202 | 0.15 |
ENSMUST00000143636.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr1_-_45542442 | 0.15 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr3_+_88624194 | 0.15 |
ENSMUST00000172252.2
|
Rit1
|
Ras-like without CAAX 1 |
| chr19_+_29902506 | 0.14 |
ENSMUST00000120388.9
ENSMUST00000144528.8 ENSMUST00000177518.8 |
Il33
|
interleukin 33 |
| chr9_-_114469121 | 0.14 |
ENSMUST00000070117.8
|
Cnot10
|
CCR4-NOT transcription complex, subunit 10 |
| chr9_+_19828161 | 0.14 |
ENSMUST00000217347.2
ENSMUST00000057596.10 |
Olfr77
|
olfactory receptor 77 |
| chr10_-_8632519 | 0.14 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr9_+_56325893 | 0.13 |
ENSMUST00000034879.5
ENSMUST00000215269.2 |
Hmg20a
|
high mobility group 20A |
| chr15_-_76491684 | 0.13 |
ENSMUST00000230157.2
|
Cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr12_+_111937978 | 0.13 |
ENSMUST00000079009.11
|
Tdrd9
|
tudor domain containing 9 |
| chr2_-_34716199 | 0.12 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr1_+_63216281 | 0.12 |
ENSMUST00000188524.2
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr13_+_16189041 | 0.11 |
ENSMUST00000164993.2
|
Inhba
|
inhibin beta-A |
| chrX_-_133012600 | 0.11 |
ENSMUST00000033610.13
|
Nox1
|
NADPH oxidase 1 |
| chr12_-_113733922 | 0.11 |
ENSMUST00000180013.3
|
Ighv2-9-1
|
immunoglobulin heavy variable 2-9-1 |
| chr10_-_40134104 | 0.11 |
ENSMUST00000217141.2
|
Gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr3_+_32490525 | 0.11 |
ENSMUST00000108242.2
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chrX_+_56257374 | 0.10 |
ENSMUST00000033466.2
|
Cd40lg
|
CD40 ligand |
| chr13_-_99653045 | 0.10 |
ENSMUST00000064762.6
|
Map1b
|
microtubule-associated protein 1B |
| chr12_+_80509978 | 0.10 |
ENSMUST00000219272.2
|
Exd2
|
exonuclease 3'-5' domain containing 2 |
| chr9_-_15191184 | 0.10 |
ENSMUST00000216406.2
|
Med17
|
mediator complex subunit 17 |
| chr10_+_89906956 | 0.10 |
ENSMUST00000183109.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_-_13871454 | 0.10 |
ENSMUST00000185219.2
ENSMUST00000185361.2 ENSMUST00000155856.2 |
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr18_+_42669322 | 0.10 |
ENSMUST00000236418.2
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr17_-_56343625 | 0.10 |
ENSMUST00000003268.11
|
Sh3gl1
|
SH3-domain GRB2-like 1 |
| chr7_+_131161951 | 0.10 |
ENSMUST00000084502.7
|
Bub3
|
BUB3 mitotic checkpoint protein |
| chr9_+_50466127 | 0.10 |
ENSMUST00000213916.2
|
Il18
|
interleukin 18 |
| chr3_+_88624145 | 0.09 |
ENSMUST00000029692.15
ENSMUST00000171645.8 |
Rit1
|
Ras-like without CAAX 1 |
| chr2_+_36575800 | 0.09 |
ENSMUST00000213258.2
|
Olfr346
|
olfactory receptor 346 |
| chr14_+_65504067 | 0.09 |
ENSMUST00000224629.2
|
Fbxo16
|
F-box protein 16 |
| chr7_+_129193581 | 0.09 |
ENSMUST00000084519.7
|
Wdr11
|
WD repeat domain 11 |
| chr17_+_79919267 | 0.09 |
ENSMUST00000223924.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr3_+_108479015 | 0.09 |
ENSMUST00000143054.2
|
Taf13
|
TATA-box binding protein associated factor 13 |
| chr14_+_73747454 | 0.09 |
ENSMUST00000022705.7
|
Med4
|
mediator complex subunit 4 |
| chr16_-_43836681 | 0.09 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chrX_-_73436293 | 0.09 |
ENSMUST00000114138.8
|
Fam3a
|
family with sequence similarity 3, member A |
| chr12_-_87435091 | 0.09 |
ENSMUST00000021424.5
|
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr4_+_150321272 | 0.09 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr1_+_163607143 | 0.09 |
ENSMUST00000077642.12
ENSMUST00000027877.7 |
Kifap3
|
kinesin-associated protein 3 |
| chr19_-_34143437 | 0.09 |
ENSMUST00000025686.9
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr18_-_36859732 | 0.08 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr19_+_34078333 | 0.08 |
ENSMUST00000025685.8
|
Lipm
|
lipase, family member M |
| chr4_+_41966058 | 0.08 |
ENSMUST00000108026.3
|
Fam205a4
|
family with sequence similarity 205, member A4 |
| chr2_+_69619991 | 0.08 |
ENSMUST00000112266.8
|
Phospho2
|
phosphatase, orphan 2 |
| chr7_-_104250951 | 0.08 |
ENSMUST00000216750.2
ENSMUST00000215538.2 |
Olfr655
|
olfactory receptor 655 |
| chr6_+_68013772 | 0.08 |
ENSMUST00000197515.2
ENSMUST00000103315.3 |
Igkv17-121
|
immunoglobulin kappa variable 17-121 |
| chr5_-_87847268 | 0.08 |
ENSMUST00000196869.5
ENSMUST00000199624.5 ENSMUST00000198057.5 ENSMUST00000082370.10 |
Csn2
|
casein beta |
| chr8_-_10027650 | 0.08 |
ENSMUST00000170033.2
|
Lig4
|
ligase IV, DNA, ATP-dependent |
| chr4_-_134495234 | 0.08 |
ENSMUST00000037828.8
|
Ldlrap1
|
low density lipoprotein receptor adaptor protein 1 |
| chr19_-_33602652 | 0.07 |
ENSMUST00000124230.3
|
Gm8978
|
predicted gene 8978 |
| chrX_-_101200670 | 0.07 |
ENSMUST00000056904.3
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 like |
| chr6_+_129510331 | 0.07 |
ENSMUST00000204956.2
ENSMUST00000204639.2 |
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr5_-_86437119 | 0.07 |
ENSMUST00000196462.2
ENSMUST00000059424.10 |
Tmprss11c
|
transmembrane protease, serine 11c |
| chr11_-_99556781 | 0.07 |
ENSMUST00000100476.3
|
Krtap4-6
|
keratin associated protein 4-6 |
| chr1_-_84818223 | 0.07 |
ENSMUST00000186465.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr2_+_110551976 | 0.07 |
ENSMUST00000090332.5
|
Muc15
|
mucin 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Emx1_Emx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.3 | 1.2 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.3 | 1.1 | GO:0017126 | nucleologenesis(GO:0017126) |
| 0.2 | 1.3 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 1.6 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 0.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.5 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.2 | 0.5 | GO:0006532 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.1 | 0.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.6 | GO:0034309 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.1 | 0.3 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.7 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.1 | 0.9 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.3 | GO:0032385 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) |
| 0.1 | 0.3 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.3 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.1 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.8 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.3 | GO:0038095 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Fc-epsilon receptor signaling pathway(GO:0038095) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.3 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0042245 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.0 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.0 | 0.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 1.2 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0043545 | molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.3 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.4 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 1.9 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.9 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:1905035 | regulation of antifungal innate immune response(GO:1905034) negative regulation of antifungal innate immune response(GO:1905035) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 1.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.4 | 1.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 0.9 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.1 | 0.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 3.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 2.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.6 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 1.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.4 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.5 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 1.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.3 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.4 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.3 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.2 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.2 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 1.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 2.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.9 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.0 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 2.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.7 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |