Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
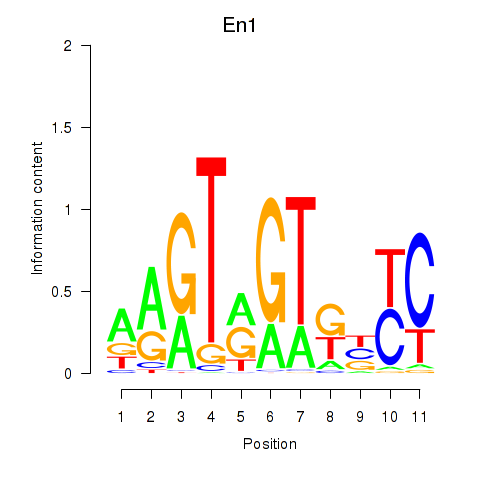
Results for En1
Z-value: 1.08
Transcription factors associated with En1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En1
|
ENSMUSG00000058665.9 | engrailed 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En1 | mm39_v1_chr1_+_120530134_120530147 | 0.42 | 4.8e-01 | Click! |
Activity profile of En1 motif
Sorted Z-values of En1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110867807 | 0.66 |
ENSMUST00000197575.2
|
Ltf
|
lactotransferrin |
| chr1_+_40554513 | 0.62 |
ENSMUST00000027237.12
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr19_-_11618165 | 0.56 |
ENSMUST00000186023.7
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr19_-_11618192 | 0.55 |
ENSMUST00000112984.4
|
Ms4a3
|
membrane-spanning 4-domains, subfamily A, member 3 |
| chr6_-_124410452 | 0.43 |
ENSMUST00000124998.2
ENSMUST00000238807.2 |
Clstn3
|
calsyntenin 3 |
| chr17_+_34124078 | 0.40 |
ENSMUST00000172817.2
|
Smim40
|
small integral membrane protein 40 |
| chr9_+_121245036 | 0.38 |
ENSMUST00000211187.2
|
Trak1
|
trafficking protein, kinesin binding 1 |
| chr19_-_10079091 | 0.37 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr2_-_71198091 | 0.35 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr2_+_72306503 | 0.34 |
ENSMUST00000102691.11
ENSMUST00000157019.2 |
Cdca7
|
cell division cycle associated 7 |
| chr3_+_89970088 | 0.34 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr19_-_32038838 | 0.32 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr19_-_40982576 | 0.31 |
ENSMUST00000117695.8
|
Blnk
|
B cell linker |
| chr14_+_71011744 | 0.29 |
ENSMUST00000022698.8
|
Dok2
|
docking protein 2 |
| chr13_-_22190575 | 0.28 |
ENSMUST00000150547.3
|
Prss16
|
protease, serine 16 (thymus) |
| chr1_-_105591362 | 0.28 |
ENSMUST00000187537.7
ENSMUST00000186485.7 ENSMUST00000190811.7 |
Pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr5_-_121045568 | 0.28 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr4_+_148125630 | 0.28 |
ENSMUST00000069604.15
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr17_+_18801989 | 0.26 |
ENSMUST00000165692.8
|
Vmn2r96
|
vomeronasal 2, receptor 96 |
| chr3_+_96543143 | 0.25 |
ENSMUST00000165842.3
|
Pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr1_+_177273226 | 0.25 |
ENSMUST00000077225.8
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr12_+_59060162 | 0.25 |
ENSMUST00000021379.8
|
Gemin2
|
gem nuclear organelle associated protein 2 |
| chr6_+_70703409 | 0.25 |
ENSMUST00000103410.3
|
Igkc
|
immunoglobulin kappa constant |
| chr17_+_36152559 | 0.24 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr17_-_44416619 | 0.24 |
ENSMUST00000143137.2
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr6_-_69415741 | 0.24 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr12_-_36206750 | 0.24 |
ENSMUST00000221388.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr11_+_87000032 | 0.22 |
ENSMUST00000020794.6
|
Ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr2_+_14234198 | 0.22 |
ENSMUST00000028045.4
|
Mrc1
|
mannose receptor, C type 1 |
| chr10_+_61010983 | 0.21 |
ENSMUST00000143207.8
ENSMUST00000148181.8 ENSMUST00000151886.8 |
Tbata
|
thymus, brain and testes associated |
| chr4_-_115980813 | 0.21 |
ENSMUST00000102704.4
ENSMUST00000102705.10 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr17_-_44416665 | 0.20 |
ENSMUST00000024757.14
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr16_+_13721016 | 0.20 |
ENSMUST00000128757.8
|
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr19_-_8691460 | 0.20 |
ENSMUST00000206560.2
ENSMUST00000205538.2 |
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr7_-_4448631 | 0.19 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr1_-_88205233 | 0.19 |
ENSMUST00000065420.12
ENSMUST00000054674.15 |
Hjurp
|
Holliday junction recognition protein |
| chrX_+_134786600 | 0.18 |
ENSMUST00000180025.8
ENSMUST00000148374.8 ENSMUST00000068755.14 |
Bhlhb9
|
basic helix-loop-helix domain containing, class B9 |
| chr5_+_137292865 | 0.18 |
ENSMUST00000052825.7
|
Ufsp1
|
UFM1-specific peptidase 1 |
| chr16_-_13720915 | 0.18 |
ENSMUST00000115803.9
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr1_+_170472092 | 0.18 |
ENSMUST00000046792.9
|
Olfml2b
|
olfactomedin-like 2B |
| chrX_-_100463810 | 0.18 |
ENSMUST00000118092.8
ENSMUST00000119699.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr8_+_89020845 | 0.17 |
ENSMUST00000098521.4
|
Adcy7
|
adenylate cyclase 7 |
| chr9_+_113641615 | 0.17 |
ENSMUST00000111838.10
ENSMUST00000166734.10 ENSMUST00000214522.2 ENSMUST00000163895.3 |
Clasp2
|
CLIP associating protein 2 |
| chr3_+_94600863 | 0.17 |
ENSMUST00000090848.10
ENSMUST00000173981.8 ENSMUST00000173849.8 ENSMUST00000174223.2 |
Selenbp2
|
selenium binding protein 2 |
| chr19_-_47003551 | 0.17 |
ENSMUST00000172239.3
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr4_+_106924181 | 0.17 |
ENSMUST00000106758.8
ENSMUST00000145324.8 ENSMUST00000106760.8 |
Cyb5rl
|
cytochrome b5 reductase-like |
| chr9_+_107468146 | 0.17 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr5_-_35886605 | 0.17 |
ENSMUST00000070203.14
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr6_-_125213911 | 0.17 |
ENSMUST00000112282.3
ENSMUST00000112281.8 ENSMUST00000032486.13 |
Cd27
|
CD27 antigen |
| chr14_-_70945434 | 0.16 |
ENSMUST00000228346.2
|
Xpo7
|
exportin 7 |
| chr7_-_24997393 | 0.16 |
ENSMUST00000005583.12
|
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr7_+_46490899 | 0.16 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chr10_-_59838815 | 0.16 |
ENSMUST00000182116.8
|
Anapc16
|
anaphase promoting complex subunit 16 |
| chr13_+_104315301 | 0.16 |
ENSMUST00000022225.12
ENSMUST00000069187.12 |
Trim23
|
tripartite motif-containing 23 |
| chr2_+_4022537 | 0.16 |
ENSMUST00000177457.8
|
Frmd4a
|
FERM domain containing 4A |
| chr4_+_152123772 | 0.16 |
ENSMUST00000084116.13
ENSMUST00000103197.5 |
Nol9
|
nucleolar protein 9 |
| chr4_-_42665763 | 0.16 |
ENSMUST00000238770.2
|
Il11ra2
|
interleukin 11 receptor, alpha chain 2 |
| chr11_-_95733235 | 0.16 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr2_+_25346841 | 0.16 |
ENSMUST00000114265.9
ENSMUST00000102918.3 |
Clic3
|
chloride intracellular channel 3 |
| chr7_-_24997291 | 0.15 |
ENSMUST00000148150.8
ENSMUST00000155118.2 |
Pafah1b3
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 |
| chr4_-_56802266 | 0.15 |
ENSMUST00000030140.3
|
Elp1
|
elongator complex protein 1 |
| chr4_-_42168603 | 0.15 |
ENSMUST00000098121.4
|
Gm13305
|
predicted gene 13305 |
| chr6_+_51447317 | 0.15 |
ENSMUST00000094623.10
|
Cbx3
|
chromobox 3 |
| chr8_-_61436249 | 0.15 |
ENSMUST00000004430.14
ENSMUST00000110301.2 ENSMUST00000093490.9 |
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr2_-_35351259 | 0.15 |
ENSMUST00000113001.9
ENSMUST00000113002.9 |
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr2_-_89239943 | 0.15 |
ENSMUST00000217226.2
ENSMUST00000217237.2 |
Olfr1238
|
olfactory receptor 1238 |
| chr2_+_30156733 | 0.15 |
ENSMUST00000113645.8
ENSMUST00000133877.8 ENSMUST00000139719.8 ENSMUST00000113643.8 ENSMUST00000150695.8 |
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr17_-_27816151 | 0.15 |
ENSMUST00000231742.2
|
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr3_-_86906591 | 0.14 |
ENSMUST00000063869.11
ENSMUST00000029717.4 |
Cd1d1
|
CD1d1 antigen |
| chr6_+_57679455 | 0.14 |
ENSMUST00000072954.8
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr2_-_69542805 | 0.14 |
ENSMUST00000102706.4
ENSMUST00000073152.13 |
Fastkd1
|
FAST kinase domains 1 |
| chr16_-_30900181 | 0.14 |
ENSMUST00000055389.9
|
Xxylt1
|
xyloside xylosyltransferase 1 |
| chr9_+_8544143 | 0.14 |
ENSMUST00000050433.8
ENSMUST00000217462.2 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr5_+_90920294 | 0.14 |
ENSMUST00000031320.8
|
Pf4
|
platelet factor 4 |
| chr17_+_28945057 | 0.14 |
ENSMUST00000233003.2
|
Gm4356
|
predicted gene 4356 |
| chr13_+_25127127 | 0.14 |
ENSMUST00000021773.13
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr19_+_44994094 | 0.14 |
ENSMUST00000236685.2
|
Twnk
|
twinkle mtDNA helicase |
| chr5_+_24791719 | 0.14 |
ENSMUST00000088295.9
ENSMUST00000121863.5 |
Chpf2
|
chondroitin polymerizing factor 2 |
| chr4_+_118721846 | 0.14 |
ENSMUST00000106360.3
ENSMUST00000216589.2 |
Olfr1331
|
olfactory receptor 1331 |
| chr8_+_72022300 | 0.13 |
ENSMUST00000212111.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr17_-_45883421 | 0.13 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr3_+_20011201 | 0.13 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr6_-_87798613 | 0.13 |
ENSMUST00000204169.2
|
Gm45140
|
predicted gene 45140 |
| chr15_-_79658608 | 0.13 |
ENSMUST00000229644.2
ENSMUST00000023055.8 |
Dnal4
|
dynein, axonemal, light chain 4 |
| chr16_+_35758836 | 0.13 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chrX_-_135072979 | 0.13 |
ENSMUST00000163584.8
ENSMUST00000060101.10 |
Tceal8
|
transcription elongation factor A (SII)-like 8 |
| chr10_-_127147609 | 0.13 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chr13_+_21295653 | 0.13 |
ENSMUST00000213326.2
ENSMUST00000215941.2 ENSMUST00000215207.2 ENSMUST00000213922.2 |
Olfr1369-ps1
|
olfactory receptor 1369, pseudogene 1 |
| chr1_-_134883645 | 0.12 |
ENSMUST00000045665.13
ENSMUST00000086444.6 ENSMUST00000112163.2 |
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr19_-_46950355 | 0.12 |
ENSMUST00000236501.2
|
Nt5c2
|
5'-nucleotidase, cytosolic II |
| chr11_+_49327451 | 0.12 |
ENSMUST00000215226.2
|
Olfr1388
|
olfactory receptor 1388 |
| chr10_+_128089965 | 0.12 |
ENSMUST00000060782.5
ENSMUST00000218722.2 |
Apon
|
apolipoprotein N |
| chr11_-_98915005 | 0.12 |
ENSMUST00000068031.8
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr16_-_26190578 | 0.12 |
ENSMUST00000023154.3
|
Cldn1
|
claudin 1 |
| chr9_+_122717536 | 0.12 |
ENSMUST00000063980.8
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr1_-_133681419 | 0.12 |
ENSMUST00000125659.8
ENSMUST00000048953.14 ENSMUST00000165602.9 |
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr1_-_63215812 | 0.12 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr17_+_26342474 | 0.12 |
ENSMUST00000025014.10
ENSMUST00000236166.2 ENSMUST00000127647.3 |
Mrpl28
|
mitochondrial ribosomal protein L28 |
| chrX_+_47235313 | 0.12 |
ENSMUST00000033427.7
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr5_+_96989853 | 0.12 |
ENSMUST00000196126.2
|
Anxa3
|
annexin A3 |
| chr12_-_108859123 | 0.12 |
ENSMUST00000161154.2
ENSMUST00000161410.8 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr5_+_31652079 | 0.12 |
ENSMUST00000076949.13
ENSMUST00000202394.4 |
Gpn1
|
GPN-loop GTPase 1 |
| chr9_+_92131797 | 0.12 |
ENSMUST00000093801.10
|
Plscr1
|
phospholipid scramblase 1 |
| chr6_+_68247469 | 0.11 |
ENSMUST00000103321.3
|
Igkv1-110
|
immunoglobulin kappa variable 1-110 |
| chr13_+_75987987 | 0.11 |
ENSMUST00000022082.8
ENSMUST00000223120.2 ENSMUST00000220523.2 |
Glrx
|
glutaredoxin |
| chrX_+_138464065 | 0.11 |
ENSMUST00000113027.8
|
Rnf128
|
ring finger protein 128 |
| chr18_-_39620115 | 0.11 |
ENSMUST00000097592.9
ENSMUST00000115571.8 |
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr9_+_81745723 | 0.11 |
ENSMUST00000057067.10
ENSMUST00000189832.7 ENSMUST00000189391.2 |
Mei4
|
meiotic double-stranded break formation protein 4 |
| chr14_+_55120777 | 0.11 |
ENSMUST00000022806.10
|
Bcl2l2
|
BCL2-like 2 |
| chr2_+_28417800 | 0.11 |
ENSMUST00000238699.2
|
Ralgds
|
ral guanine nucleotide dissociation stimulator |
| chr2_-_91274967 | 0.11 |
ENSMUST00000064652.14
ENSMUST00000102594.11 ENSMUST00000094835.9 |
1110051M20Rik
|
RIKEN cDNA 1110051M20 gene |
| chr7_-_34755985 | 0.11 |
ENSMUST00000130491.3
|
Cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr10_+_116013256 | 0.11 |
ENSMUST00000155606.8
ENSMUST00000128399.2 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr13_-_48746836 | 0.11 |
ENSMUST00000238995.2
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chrX_-_100463395 | 0.11 |
ENSMUST00000117901.8
ENSMUST00000120201.8 ENSMUST00000117637.8 ENSMUST00000134005.2 ENSMUST00000121520.8 |
Zmym3
|
zinc finger, MYM-type 3 |
| chr4_-_106656976 | 0.11 |
ENSMUST00000145061.8
ENSMUST00000102762.10 |
Acot11
|
acyl-CoA thioesterase 11 |
| chr9_+_108820846 | 0.11 |
ENSMUST00000198140.5
ENSMUST00000051873.15 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr7_-_80020830 | 0.10 |
ENSMUST00000205436.2
ENSMUST00000098346.5 |
Man2a2
|
mannosidase 2, alpha 2 |
| chr6_-_56878854 | 0.10 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr1_+_88154727 | 0.10 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr17_+_34416689 | 0.10 |
ENSMUST00000173441.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr9_+_64024429 | 0.10 |
ENSMUST00000034969.14
|
Lctl
|
lactase-like |
| chr7_-_126399208 | 0.10 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr10_-_8632519 | 0.10 |
ENSMUST00000212869.2
|
Sash1
|
SAM and SH3 domain containing 1 |
| chr1_-_191307648 | 0.10 |
ENSMUST00000027933.11
|
Dtl
|
denticleless E3 ubiquitin protein ligase |
| chr6_+_92069376 | 0.10 |
ENSMUST00000113463.8
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr3_+_152101080 | 0.10 |
ENSMUST00000106103.8
|
Zzz3
|
zinc finger, ZZ domain containing 3 |
| chr11_+_75239259 | 0.10 |
ENSMUST00000044530.3
|
Smyd4
|
SET and MYND domain containing 4 |
| chr8_-_27618643 | 0.10 |
ENSMUST00000033877.6
|
Brf2
|
BRF2, RNA polymerase III transcription initiation factor 50kDa subunit |
| chr12_+_117480099 | 0.10 |
ENSMUST00000109691.4
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_-_35332101 | 0.10 |
ENSMUST00000131745.8
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr5_+_137628377 | 0.10 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr5_-_110927803 | 0.10 |
ENSMUST00000112426.8
|
Pus1
|
pseudouridine synthase 1 |
| chr6_-_87786736 | 0.10 |
ENSMUST00000032134.9
|
Rab43
|
RAB43, member RAS oncogene family |
| chr10_-_56104732 | 0.09 |
ENSMUST00000099739.5
|
Tbc1d32
|
TBC1 domain family, member 32 |
| chr2_-_11558703 | 0.09 |
ENSMUST00000192949.6
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr5_+_31079177 | 0.09 |
ENSMUST00000031053.15
ENSMUST00000202752.2 |
Khk
|
ketohexokinase |
| chr4_-_149760488 | 0.09 |
ENSMUST00000118704.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr6_+_29859372 | 0.09 |
ENSMUST00000115238.10
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr16_+_38279289 | 0.09 |
ENSMUST00000099816.3
ENSMUST00000232409.2 |
Cd80
|
CD80 antigen |
| chr14_-_33996185 | 0.09 |
ENSMUST00000227006.2
|
Shld2
|
shieldin complex subunit 2 |
| chr17_-_23990512 | 0.09 |
ENSMUST00000226460.2
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr15_-_79658584 | 0.09 |
ENSMUST00000069877.12
|
Dnal4
|
dynein, axonemal, light chain 4 |
| chr12_-_36206626 | 0.09 |
ENSMUST00000220828.2
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr17_-_31417834 | 0.09 |
ENSMUST00000236793.2
|
Tmprss3
|
transmembrane protease, serine 3 |
| chr14_-_70405288 | 0.09 |
ENSMUST00000129174.8
ENSMUST00000125300.3 |
Pdlim2
|
PDZ and LIM domain 2 |
| chr9_-_119813724 | 0.09 |
ENSMUST00000213936.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr6_-_50374048 | 0.09 |
ENSMUST00000136926.3
|
Osbpl3
|
oxysterol binding protein-like 3 |
| chr7_+_30264835 | 0.09 |
ENSMUST00000043850.14
|
Igflr1
|
IGF-like family receptor 1 |
| chrX_+_20570145 | 0.09 |
ENSMUST00000033383.3
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr6_+_68402550 | 0.09 |
ENSMUST00000103323.3
|
Igkv16-104
|
immunoglobulin kappa variable 16-104 |
| chr19_-_8775935 | 0.09 |
ENSMUST00000096261.5
|
Polr2g
|
polymerase (RNA) II (DNA directed) polypeptide G |
| chr12_+_76353835 | 0.09 |
ENSMUST00000220321.2
|
Mthfd1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase |
| chr12_+_112940361 | 0.09 |
ENSMUST00000223368.2
ENSMUST00000223508.2 |
Btbd6
|
BTB (POZ) domain containing 6 |
| chr7_-_121306476 | 0.09 |
ENSMUST00000046929.7
|
Usp31
|
ubiquitin specific peptidase 31 |
| chr7_-_126194097 | 0.09 |
ENSMUST00000058429.6
|
Il27
|
interleukin 27 |
| chr14_-_62998561 | 0.09 |
ENSMUST00000053959.7
ENSMUST00000223585.2 |
Ints6
|
integrator complex subunit 6 |
| chr9_-_45847344 | 0.09 |
ENSMUST00000034590.4
|
Tagln
|
transgelin |
| chr17_-_75858835 | 0.09 |
ENSMUST00000234785.2
ENSMUST00000112507.4 |
Fam98a
|
family with sequence similarity 98, member A |
| chr3_-_14843512 | 0.09 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr4_+_115685257 | 0.09 |
ENSMUST00000030477.4
|
Mob3c
|
MOB kinase activator 3C |
| chr7_+_83281167 | 0.08 |
ENSMUST00000075418.15
|
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr17_+_34416707 | 0.08 |
ENSMUST00000025196.9
|
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr4_-_123611974 | 0.08 |
ENSMUST00000137312.2
ENSMUST00000106206.8 |
Ndufs5
|
NADH:ubiquinone oxidoreductase core subunit S5 |
| chrX_-_56326951 | 0.08 |
ENSMUST00000114735.9
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr9_+_65494469 | 0.08 |
ENSMUST00000239405.2
ENSMUST00000047099.13 ENSMUST00000131483.3 ENSMUST00000141046.3 |
Pif1
|
PIF1 5'-to-3' DNA helicase |
| chr6_+_86381201 | 0.08 |
ENSMUST00000095754.10
ENSMUST00000095753.9 |
Tia1
|
cytotoxic granule-associated RNA binding protein 1 |
| chrX_+_135950334 | 0.08 |
ENSMUST00000047852.8
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr10_-_25076008 | 0.08 |
ENSMUST00000100012.3
|
Akap7
|
A kinase (PRKA) anchor protein 7 |
| chr9_+_111011327 | 0.08 |
ENSMUST00000216430.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr9_+_111011388 | 0.08 |
ENSMUST00000217117.2
|
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chrX_-_55643429 | 0.08 |
ENSMUST00000059899.3
|
Mmgt1
|
membrane magnesium transporter 1 |
| chr9_+_54858388 | 0.08 |
ENSMUST00000171900.2
|
Psma4
|
proteasome subunit alpha 4 |
| chr6_-_69204417 | 0.08 |
ENSMUST00000103346.3
|
Igkv4-72
|
immunoglobulin kappa chain variable 4-72 |
| chr12_+_98234884 | 0.08 |
ENSMUST00000075072.6
|
Gpr65
|
G-protein coupled receptor 65 |
| chr16_-_13720949 | 0.08 |
ENSMUST00000023361.12
ENSMUST00000115802.2 |
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr14_+_55120875 | 0.08 |
ENSMUST00000134077.2
ENSMUST00000172844.8 ENSMUST00000133397.4 ENSMUST00000227108.2 |
Gm20521
Bcl2l2
|
predicted gene 20521 BCL2-like 2 |
| chr7_+_51537645 | 0.08 |
ENSMUST00000208711.2
|
Gas2
|
growth arrest specific 2 |
| chr15_-_78056618 | 0.08 |
ENSMUST00000229476.2
|
Ift27
|
intraflagellar transport 27 |
| chr2_-_155199300 | 0.07 |
ENSMUST00000165234.2
ENSMUST00000077626.13 |
Pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr8_+_124750133 | 0.07 |
ENSMUST00000034457.9
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr4_-_44066960 | 0.07 |
ENSMUST00000173234.8
ENSMUST00000173274.2 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr7_+_125307060 | 0.07 |
ENSMUST00000124223.8
ENSMUST00000069660.13 |
Katnip
|
katanin interacting protein |
| chr19_-_11243530 | 0.07 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr16_+_20367327 | 0.07 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr3_-_96812610 | 0.07 |
ENSMUST00000029738.14
|
Gpr89
|
G protein-coupled receptor 89 |
| chr4_+_122910382 | 0.07 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr11_+_72687080 | 0.07 |
ENSMUST00000207107.2
|
Zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr10_+_4382467 | 0.07 |
ENSMUST00000095893.11
ENSMUST00000118544.8 ENSMUST00000117489.8 |
Armt1
|
acidic residue methyltransferase 1 |
| chr1_+_163875783 | 0.07 |
ENSMUST00000027874.6
|
Sele
|
selectin, endothelial cell |
| chr12_+_102710008 | 0.07 |
ENSMUST00000057416.8
|
Tmem251
|
transmembrane protein 251 |
| chr13_-_49473695 | 0.07 |
ENSMUST00000110086.2
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr6_+_90179768 | 0.07 |
ENSMUST00000078371.6
|
V1ra8
|
vomeronasal 1 receptor, A8 |
| chr10_-_18890281 | 0.07 |
ENSMUST00000146388.2
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chrX_+_139857640 | 0.07 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr1_+_105591595 | 0.07 |
ENSMUST00000039173.13
ENSMUST00000086721.10 ENSMUST00000190501.7 |
Relch
|
RAB11 binding and LisH domain, coiled-coil and HEAT repeat containing |
| chr16_-_45830575 | 0.07 |
ENSMUST00000130481.2
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr17_-_25105277 | 0.06 |
ENSMUST00000234583.2
ENSMUST00000234968.2 ENSMUST00000044252.7 |
Nubp2
|
nucleotide binding protein 2 |
| chr9_-_44891626 | 0.06 |
ENSMUST00000002101.12
ENSMUST00000160886.2 |
Cd3g
|
CD3 antigen, gamma polypeptide |
| chr16_-_55755160 | 0.06 |
ENSMUST00000122280.8
ENSMUST00000121703.3 |
Cep97
|
centrosomal protein 97 |
Network of associatons between targets according to the STRING database.
First level regulatory network of En1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:0033580 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 | 0.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:0048003 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:1903660 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0006507 | GPI anchor release(GO:0006507) regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.2 | GO:0035660 | MyD88-dependent toll-like receptor 4 signaling pathway(GO:0035660) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0002358 | B cell homeostatic proliferation(GO:0002358) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0034148 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 | 0.1 | GO:0009257 | histidine biosynthetic process(GO:0000105) 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:2000582 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:1904826 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 0.2 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.0 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.0 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.0 | GO:0030845 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:2000156 | regulation of retrograde vesicle-mediated transport, Golgi to ER(GO:2000156) |
| 0.0 | 0.1 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.2 | GO:1904511 | cortical microtubule plus-end(GO:1903754) cytoplasmic microtubule plus-end(GO:1904511) |
| 0.0 | 0.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.6 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.3 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.0 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0004454 | ketohexokinase activity(GO:0004454) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.1 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |