Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
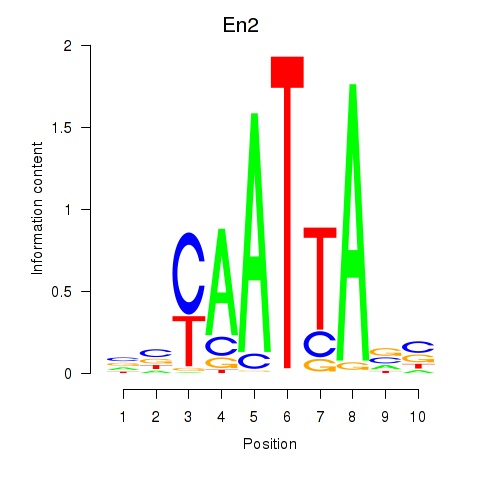
Results for En2
Z-value: 0.96
Transcription factors associated with En2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En2
|
ENSMUSG00000039095.9 | engrailed 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| En2 | mm39_v1_chr5_+_28370687_28370720 | -0.09 | 8.9e-01 | Click! |
Activity profile of En2 motif
Sorted Z-values of En2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23728222 | 2.86 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7 |
| chr3_+_96175970 | 2.13 |
ENSMUST00000098843.3
|
H3c13
|
H3 clustered histone 13 |
| chr13_+_21901791 | 1.73 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10 |
| chr13_-_22017677 | 1.09 |
ENSMUST00000081342.7
|
H2ac24
|
H2A clustered histone 24 |
| chr13_+_23758555 | 1.00 |
ENSMUST00000090776.7
|
H2ac7
|
H2A clustered histone 7 |
| chrM_+_11735 | 0.78 |
ENSMUST00000082418.1
|
mt-Nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chrM_+_9870 | 0.76 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chrM_+_3906 | 0.72 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr12_+_108145802 | 0.64 |
ENSMUST00000221167.2
|
Ccnk
|
cyclin K |
| chr13_-_22227114 | 0.63 |
ENSMUST00000091741.6
|
H2ac11
|
H2A clustered histone 11 |
| chrM_+_2743 | 0.49 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr7_-_25454177 | 0.45 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr9_-_107512566 | 0.44 |
ENSMUST00000055704.12
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chrM_+_7006 | 0.43 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr1_-_92412835 | 0.42 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416 |
| chr2_-_111843053 | 0.42 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310 |
| chr4_+_42949814 | 0.41 |
ENSMUST00000037872.10
ENSMUST00000098112.9 |
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr12_+_108145997 | 0.39 |
ENSMUST00000101055.5
|
Ccnk
|
cyclin K |
| chr8_-_32440071 | 0.38 |
ENSMUST00000207678.3
|
Nrg1
|
neuregulin 1 |
| chrM_+_10167 | 0.38 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chrX_+_101163053 | 0.37 |
ENSMUST00000113627.4
|
Pin4
|
protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr5_-_124387812 | 0.36 |
ENSMUST00000162812.8
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chrX_+_128650486 | 0.35 |
ENSMUST00000167619.9
ENSMUST00000037854.15 |
Diaph2
|
diaphanous related formin 2 |
| chr16_+_33504740 | 0.35 |
ENSMUST00000232568.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr9_-_107512511 | 0.35 |
ENSMUST00000192615.6
ENSMUST00000192837.2 ENSMUST00000193876.2 |
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr7_-_25454126 | 0.34 |
ENSMUST00000108401.3
ENSMUST00000043765.14 |
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr18_+_39126325 | 0.31 |
ENSMUST00000137497.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr8_-_84321032 | 0.31 |
ENSMUST00000163837.2
|
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr9_+_20193647 | 0.30 |
ENSMUST00000071725.4
ENSMUST00000212983.3 |
Olfr39
|
olfactory receptor 39 |
| chr8_-_84321069 | 0.29 |
ENSMUST00000019382.17
ENSMUST00000212630.2 ENSMUST00000165740.9 |
Tecr
|
trans-2,3-enoyl-CoA reductase |
| chr19_+_6096606 | 0.29 |
ENSMUST00000138532.8
ENSMUST00000129081.8 ENSMUST00000156550.8 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr18_+_39126178 | 0.28 |
ENSMUST00000097593.9
|
Arhgap26
|
Rho GTPase activating protein 26 |
| chr5_+_135135735 | 0.28 |
ENSMUST00000201977.4
ENSMUST00000005507.10 |
Mlxipl
|
MLX interacting protein-like |
| chr18_+_42644552 | 0.27 |
ENSMUST00000237602.2
ENSMUST00000236088.2 ENSMUST00000025375.15 |
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr11_-_109502243 | 0.26 |
ENSMUST00000103060.10
ENSMUST00000047186.10 ENSMUST00000106689.2 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr7_-_30259253 | 0.26 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr7_-_30259025 | 0.26 |
ENSMUST00000043975.11
ENSMUST00000156241.2 |
Lin37
|
lin-37 homolog (C. elegans) |
| chr5_-_84565218 | 0.25 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5 |
| chr5_-_106844396 | 0.23 |
ENSMUST00000137285.8
ENSMUST00000124263.2 ENSMUST00000112695.4 ENSMUST00000155495.8 ENSMUST00000135108.2 ENSMUST00000149128.3 |
Zfp644
Gm28039
|
zinc finger protein 644 predicted gene, 28039 |
| chr5_-_143846600 | 0.23 |
ENSMUST00000031613.11
ENSMUST00000100483.3 |
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr1_-_155848917 | 0.23 |
ENSMUST00000138762.8
|
Cep350
|
centrosomal protein 350 |
| chr19_-_4171536 | 0.23 |
ENSMUST00000025767.14
|
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr2_-_153079828 | 0.22 |
ENSMUST00000109795.2
|
Plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr16_+_33504908 | 0.22 |
ENSMUST00000126532.2
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr11_+_68986043 | 0.22 |
ENSMUST00000101004.9
|
Per1
|
period circadian clock 1 |
| chr12_-_85197985 | 0.22 |
ENSMUST00000019379.9
ENSMUST00000221972.2 |
Rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr2_-_45007407 | 0.22 |
ENSMUST00000176438.9
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr14_+_58308004 | 0.21 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr1_+_92545510 | 0.21 |
ENSMUST00000213247.2
|
Olfr12
|
olfactory receptor 12 |
| chr10_-_12745109 | 0.21 |
ENSMUST00000218635.2
|
Utrn
|
utrophin |
| chr2_+_152596075 | 0.21 |
ENSMUST00000010020.12
|
Cox4i2
|
cytochrome c oxidase subunit 4I2 |
| chr3_-_146388165 | 0.20 |
ENSMUST00000124931.8
ENSMUST00000147113.2 |
Samd13
|
sterile alpha motif domain containing 13 |
| chr5_-_131645437 | 0.20 |
ENSMUST00000161804.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr2_-_90054837 | 0.20 |
ENSMUST00000213994.3
|
Olfr1506
|
olfactory receptor 1506 |
| chrX_-_156198282 | 0.19 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_+_136038263 | 0.19 |
ENSMUST00000105850.8
ENSMUST00000148843.10 |
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr8_-_3675525 | 0.19 |
ENSMUST00000144977.2
ENSMUST00000136105.8 ENSMUST00000128566.8 |
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr4_+_136038301 | 0.19 |
ENSMUST00000084219.12
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr4_+_136038243 | 0.18 |
ENSMUST00000131671.8
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr2_+_90229027 | 0.18 |
ENSMUST00000216111.3
|
Olfr1274
|
olfactory receptor 1274 |
| chr9_+_19716202 | 0.18 |
ENSMUST00000212540.3
ENSMUST00000217280.2 |
Olfr859
|
olfactory receptor 859 |
| chr1_-_131455039 | 0.17 |
ENSMUST00000097588.9
ENSMUST00000186543.7 |
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr5_-_106844685 | 0.17 |
ENSMUST00000127434.8
ENSMUST00000112696.8 ENSMUST00000112698.8 |
Zfp644
|
zinc finger protein 644 |
| chr5_-_62923463 | 0.17 |
ENSMUST00000076623.8
ENSMUST00000159470.3 |
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr17_-_47813201 | 0.17 |
ENSMUST00000233174.2
ENSMUST00000233121.2 ENSMUST00000067103.4 |
Taf8
|
TATA-box binding protein associated factor 8 |
| chr17_-_29226886 | 0.17 |
ENSMUST00000232723.2
|
Stk38
|
serine/threonine kinase 38 |
| chr14_-_70666513 | 0.17 |
ENSMUST00000226426.2
ENSMUST00000048129.6 |
Piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr6_-_116693849 | 0.16 |
ENSMUST00000056623.13
|
Tmem72
|
transmembrane protein 72 |
| chr5_+_36641922 | 0.16 |
ENSMUST00000060100.3
|
Ccdc96
|
coiled-coil domain containing 96 |
| chr16_+_37909363 | 0.16 |
ENSMUST00000023507.13
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chrX_+_23559282 | 0.16 |
ENSMUST00000035766.13
ENSMUST00000101670.3 |
Wdr44
|
WD repeat domain 44 |
| chr16_+_3648742 | 0.15 |
ENSMUST00000214238.2
ENSMUST00000214590.2 |
Olfr15
|
olfactory receptor 15 |
| chr19_-_12100560 | 0.15 |
ENSMUST00000213471.2
ENSMUST00000214676.2 |
Olfr76
|
olfactory receptor 76 |
| chr11_-_86884507 | 0.14 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr12_-_85327136 | 0.14 |
ENSMUST00000065913.8
ENSMUST00000008966.13 |
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr10_+_85222677 | 0.14 |
ENSMUST00000105307.8
ENSMUST00000020231.10 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr7_+_25380263 | 0.13 |
ENSMUST00000205325.2
ENSMUST00000206913.2 |
B9d2
|
B9 protein domain 2 |
| chr7_+_63835285 | 0.13 |
ENSMUST00000206263.2
ENSMUST00000206107.2 ENSMUST00000205731.2 ENSMUST00000206706.2 ENSMUST00000205690.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr10_-_128918779 | 0.12 |
ENSMUST00000213579.2
|
Olfr767
|
olfactory receptor 767 |
| chr17_+_34457868 | 0.12 |
ENSMUST00000095342.11
ENSMUST00000167280.8 ENSMUST00000236838.2 |
H2-Ob
|
histocompatibility 2, O region beta locus |
| chr1_+_104696235 | 0.11 |
ENSMUST00000062528.9
|
Cdh20
|
cadherin 20 |
| chr19_+_12364643 | 0.11 |
ENSMUST00000217062.3
ENSMUST00000216145.2 ENSMUST00000213657.2 |
Olfr1440
|
olfactory receptor 1440 |
| chrX_+_20554193 | 0.11 |
ENSMUST00000115364.8
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr8_-_84963653 | 0.11 |
ENSMUST00000039480.7
|
Zswim4
|
zinc finger SWIM-type containing 4 |
| chr11_+_75422516 | 0.11 |
ENSMUST00000149727.8
ENSMUST00000108433.8 ENSMUST00000042561.14 ENSMUST00000143035.8 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr4_+_108576846 | 0.10 |
ENSMUST00000178992.2
|
3110021N24Rik
|
RIKEN cDNA 3110021N24 gene |
| chr9_-_96601574 | 0.10 |
ENSMUST00000128269.8
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr15_+_82230155 | 0.10 |
ENSMUST00000023086.15
|
Smdt1
|
single-pass membrane protein with aspartate rich tail 1 |
| chr15_+_102314578 | 0.09 |
ENSMUST00000170884.8
ENSMUST00000163709.8 |
Sp1
|
trans-acting transcription factor 1 |
| chrX_+_159551009 | 0.09 |
ENSMUST00000033650.14
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human) |
| chr17_-_37523969 | 0.09 |
ENSMUST00000060728.7
ENSMUST00000216318.2 |
Olfr95
|
olfactory receptor 95 |
| chr18_+_82932747 | 0.09 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr10_-_44024843 | 0.09 |
ENSMUST00000200401.2
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr17_+_21165573 | 0.09 |
ENSMUST00000007708.14
|
Ppp2r1a
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr13_+_75855695 | 0.08 |
ENSMUST00000222194.2
ENSMUST00000223535.2 ENSMUST00000222853.2 |
Ell2
|
elongation factor for RNA polymerase II 2 |
| chr10_+_73657689 | 0.08 |
ENSMUST00000064562.14
ENSMUST00000193174.6 ENSMUST00000105426.10 ENSMUST00000129404.9 ENSMUST00000131321.9 ENSMUST00000126920.9 ENSMUST00000147189.9 ENSMUST00000105424.10 ENSMUST00000092420.13 ENSMUST00000105429.10 ENSMUST00000193361.6 ENSMUST00000131724.9 ENSMUST00000152655.9 ENSMUST00000151116.9 ENSMUST00000155701.9 ENSMUST00000152819.9 ENSMUST00000125517.9 ENSMUST00000124046.8 ENSMUST00000146682.8 ENSMUST00000177107.8 ENSMUST00000149977.9 ENSMUST00000191854.6 |
Pcdh15
|
protocadherin 15 |
| chr17_+_21031817 | 0.08 |
ENSMUST00000232810.2
ENSMUST00000233712.2 ENSMUST00000232852.2 |
Vmn1r229
|
vomeronasal 1 receptor 229 |
| chr8_-_108129829 | 0.08 |
ENSMUST00000003947.9
|
Nqo1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr17_-_29226965 | 0.08 |
ENSMUST00000009138.13
ENSMUST00000119274.3 |
Stk38
|
serine/threonine kinase 38 |
| chr12_+_52746158 | 0.08 |
ENSMUST00000095737.5
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr11_+_68950533 | 0.08 |
ENSMUST00000051888.4
|
Borcs6
|
BLOC-1 related complex subunit 6 |
| chr5_-_36641456 | 0.07 |
ENSMUST00000119916.2
ENSMUST00000031097.8 |
Tada2b
|
transcriptional adaptor 2B |
| chr3_+_103740056 | 0.07 |
ENSMUST00000106822.2
|
Bcl2l15
|
BCLl2-like 15 |
| chr1_-_92446383 | 0.07 |
ENSMUST00000062353.12
|
Olfr1414
|
olfactory receptor 1414 |
| chr9_-_71070506 | 0.07 |
ENSMUST00000074465.9
|
Aqp9
|
aquaporin 9 |
| chr6_-_30304512 | 0.07 |
ENSMUST00000094543.3
ENSMUST00000102993.10 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr17_-_29226700 | 0.07 |
ENSMUST00000233441.2
|
Stk38
|
serine/threonine kinase 38 |
| chr14_+_32321341 | 0.07 |
ENSMUST00000187377.7
ENSMUST00000189022.8 ENSMUST00000186452.7 |
Prrxl1
|
paired related homeobox protein-like 1 |
| chr11_+_75422953 | 0.07 |
ENSMUST00000127226.3
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr16_+_44913974 | 0.07 |
ENSMUST00000099498.10
|
Ccdc80
|
coiled-coil domain containing 80 |
| chrM_+_9459 | 0.07 |
ENSMUST00000082411.1
|
mt-Nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr3_+_75464837 | 0.06 |
ENSMUST00000161776.8
ENSMUST00000029423.9 |
Serpini1
|
serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| chr14_+_60615128 | 0.06 |
ENSMUST00000022561.9
|
Amer2
|
APC membrane recruitment 2 |
| chr7_+_106630381 | 0.06 |
ENSMUST00000213623.2
|
Olfr713
|
olfactory receptor 713 |
| chr17_+_3447465 | 0.06 |
ENSMUST00000072156.7
|
Tiam2
|
T cell lymphoma invasion and metastasis 2 |
| chr12_+_102094977 | 0.06 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr8_+_84682136 | 0.06 |
ENSMUST00000005607.9
|
Asf1b
|
anti-silencing function 1B histone chaperone |
| chr9_+_20209828 | 0.06 |
ENSMUST00000215540.2
ENSMUST00000075717.7 |
Olfr873
|
olfactory receptor 873 |
| chr13_+_118851214 | 0.05 |
ENSMUST00000022246.9
|
Fgf10
|
fibroblast growth factor 10 |
| chr7_+_78922947 | 0.05 |
ENSMUST00000037315.13
|
Abhd2
|
abhydrolase domain containing 2 |
| chr2_+_89821818 | 0.05 |
ENSMUST00000216953.3
|
Olfr1261
|
olfactory receptor 1261 |
| chr15_+_39522905 | 0.05 |
ENSMUST00000226410.2
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr2_-_84545504 | 0.05 |
ENSMUST00000035840.6
|
Zdhhc5
|
zinc finger, DHHC domain containing 5 |
| chr11_-_73290321 | 0.05 |
ENSMUST00000131253.2
ENSMUST00000120303.9 |
Olfr1
|
olfactory receptor 1 |
| chr11_+_75422925 | 0.05 |
ENSMUST00000169547.9
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr11_-_118103540 | 0.05 |
ENSMUST00000106302.9
ENSMUST00000151165.2 |
Cyth1
|
cytohesin 1 |
| chr12_-_111780268 | 0.05 |
ENSMUST00000021715.6
|
Xrcc3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr2_+_89821565 | 0.04 |
ENSMUST00000111509.3
ENSMUST00000213909.3 |
Olfr1261
|
olfactory receptor 1261 |
| chr3_+_96537235 | 0.04 |
ENSMUST00000048915.11
ENSMUST00000196456.5 ENSMUST00000198027.5 |
Rbm8a
|
RNA binding motif protein 8a |
| chr17_+_34544632 | 0.04 |
ENSMUST00000050325.10
|
H2-Eb2
|
histocompatibility 2, class II antigen E beta2 |
| chr11_+_120358461 | 0.04 |
ENSMUST00000140862.7
ENSMUST00000106205.9 ENSMUST00000106203.9 |
Hgs
|
HGF-regulated tyrosine kinase substrate |
| chr9_+_32027335 | 0.04 |
ENSMUST00000174641.8
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr10_-_126956991 | 0.04 |
ENSMUST00000080975.6
ENSMUST00000164259.9 |
Os9
|
amplified in osteosarcoma |
| chr5_+_138185747 | 0.04 |
ENSMUST00000110934.9
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr2_-_27365633 | 0.04 |
ENSMUST00000138693.8
ENSMUST00000113941.9 ENSMUST00000077737.13 |
Brd3
|
bromodomain containing 3 |
| chr11_-_69127848 | 0.04 |
ENSMUST00000021259.9
ENSMUST00000108665.8 ENSMUST00000108664.2 |
Gucy2e
|
guanylate cyclase 2e |
| chr2_-_57004933 | 0.04 |
ENSMUST00000028166.9
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr19_-_28945194 | 0.03 |
ENSMUST00000162110.8
|
Spata6l
|
spermatogenesis associated 6 like |
| chr6_+_37847721 | 0.03 |
ENSMUST00000031859.14
ENSMUST00000120428.8 |
Trim24
|
tripartite motif-containing 24 |
| chr5_-_143279378 | 0.03 |
ENSMUST00000212715.2
|
Zfp853
|
zinc finger protein 853 |
| chr8_-_45835234 | 0.03 |
ENSMUST00000093526.13
|
Fam149a
|
family with sequence similarity 149, member A |
| chr11_+_116734104 | 0.03 |
ENSMUST00000106370.10
|
Mettl23
|
methyltransferase like 23 |
| chr5_-_146158201 | 0.03 |
ENSMUST00000161574.8
|
Rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr7_+_141503411 | 0.03 |
ENSMUST00000078200.12
ENSMUST00000018971.15 |
Brsk2
|
BR serine/threonine kinase 2 |
| chr2_+_177760768 | 0.03 |
ENSMUST00000108917.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr12_-_84497718 | 0.03 |
ENSMUST00000085192.7
ENSMUST00000220491.2 |
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr4_+_21848039 | 0.03 |
ENSMUST00000098238.9
ENSMUST00000108229.2 |
Pnisr
|
PNN interacting serine/arginine-rich |
| chr11_+_95915366 | 0.03 |
ENSMUST00000103157.10
|
Gip
|
gastric inhibitory polypeptide |
| chr19_-_56810593 | 0.03 |
ENSMUST00000118592.8
|
Ccdc186
|
coiled-coil domain containing 186 |
| chr1_+_127657142 | 0.03 |
ENSMUST00000038006.8
|
Acmsd
|
amino carboxymuconate semialdehyde decarboxylase |
| chr1_-_132318039 | 0.02 |
ENSMUST00000132435.2
|
Tmcc2
|
transmembrane and coiled-coil domains 2 |
| chr16_+_33504829 | 0.02 |
ENSMUST00000152782.8
|
Heg1
|
heart development protein with EGF-like domains 1 |
| chr2_-_122016670 | 0.02 |
ENSMUST00000028665.5
|
Patl2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr18_+_34757666 | 0.02 |
ENSMUST00000167161.9
|
Kif20a
|
kinesin family member 20A |
| chr5_-_53864874 | 0.02 |
ENSMUST00000031093.5
|
Cckar
|
cholecystokinin A receptor |
| chr10_-_128361731 | 0.02 |
ENSMUST00000026427.8
|
Esyt1
|
extended synaptotagmin-like protein 1 |
| chr4_+_109835224 | 0.02 |
ENSMUST00000061187.4
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr14_+_33662976 | 0.02 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr14_+_50360643 | 0.02 |
ENSMUST00000215317.2
|
Olfr727
|
olfactory receptor 727 |
| chr18_+_84869456 | 0.02 |
ENSMUST00000160180.9
|
Cyb5a
|
cytochrome b5 type A (microsomal) |
| chr3_-_32546380 | 0.02 |
ENSMUST00000164954.3
|
Kcnmb3
|
potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| chr12_-_56660054 | 0.02 |
ENSMUST00000072631.6
|
Nkx2-9
|
NK2 homeobox 9 |
| chr11_-_99265721 | 0.02 |
ENSMUST00000006963.3
|
Krt28
|
keratin 28 |
| chr4_-_43823866 | 0.02 |
ENSMUST00000215406.2
ENSMUST00000079234.6 ENSMUST00000214843.2 |
Olfr156
|
olfactory receptor 156 |
| chr8_-_3675024 | 0.01 |
ENSMUST00000133459.8
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr3_-_151953894 | 0.01 |
ENSMUST00000196529.5
|
Nexn
|
nexilin |
| chr6_+_42762027 | 0.01 |
ENSMUST00000217701.2
ENSMUST00000215719.2 ENSMUST00000214465.2 |
Olfr452
|
olfactory receptor 452 |
| chr3_+_113824181 | 0.01 |
ENSMUST00000123619.8
ENSMUST00000092155.12 |
Col11a1
|
collagen, type XI, alpha 1 |
| chr8_+_21382681 | 0.01 |
ENSMUST00000098908.4
|
Defb33
|
defensin beta 33 |
| chr1_+_173924457 | 0.01 |
ENSMUST00000213832.2
|
Olfr427
|
olfactory receptor 427 |
| chr12_+_99930757 | 0.01 |
ENSMUST00000160413.8
ENSMUST00000177549.8 ENSMUST00000049788.9 |
Kcnk13
|
potassium channel, subfamily K, member 13 |
| chr10_+_127734384 | 0.01 |
ENSMUST00000047134.8
|
Sdr9c7
|
4short chain dehydrogenase/reductase family 9C, member 7 |
| chr18_+_23548534 | 0.01 |
ENSMUST00000221880.2
ENSMUST00000220904.2 ENSMUST00000047954.15 |
Dtna
|
dystrobrevin alpha |
| chr2_-_111854754 | 0.01 |
ENSMUST00000213602.2
ENSMUST00000215321.2 |
Olfr1311
|
olfactory receptor 1311 |
| chr10_+_88721854 | 0.01 |
ENSMUST00000020255.8
|
Slc5a8
|
solute carrier family 5 (iodide transporter), member 8 |
| chr18_-_88945571 | 0.01 |
ENSMUST00000147313.2
|
Socs6
|
suppressor of cytokine signaling 6 |
| chr11_+_114742331 | 0.01 |
ENSMUST00000177952.8
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr16_-_45544960 | 0.01 |
ENSMUST00000096057.5
|
Tagln3
|
transgelin 3 |
| chr13_-_22289994 | 0.01 |
ENSMUST00000227357.2
ENSMUST00000228428.2 |
Vmn1r189
|
vomeronasal 1 receptor 189 |
| chr2_+_153780138 | 0.01 |
ENSMUST00000109757.8
ENSMUST00000154281.3 |
Bpifb4
|
BPI fold containing family B, member 4 |
| chr8_+_95807814 | 0.01 |
ENSMUST00000034239.9
|
Katnb1
|
katanin p80 (WD40-containing) subunit B 1 |
| chr7_+_44240310 | 0.01 |
ENSMUST00000107906.6
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3 |
| chr11_+_114741948 | 0.01 |
ENSMUST00000133245.2
ENSMUST00000122967.3 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr8_+_121842902 | 0.01 |
ENSMUST00000054691.8
|
Foxc2
|
forkhead box C2 |
| chr15_+_100262423 | 0.01 |
ENSMUST00000175683.8
ENSMUST00000177211.2 |
Higd1c
|
HIG1 domain family, member 1C |
| chr9_+_43222104 | 0.01 |
ENSMUST00000034511.7
|
Trim29
|
tripartite motif-containing 29 |
| chr16_+_3410262 | 0.01 |
ENSMUST00000061541.2
|
Olfr161
|
olfactory receptor 161 |
| chr3_+_63203516 | 0.01 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr17_-_36008863 | 0.01 |
ENSMUST00000146472.8
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr8_+_82582953 | 0.01 |
ENSMUST00000109851.3
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr1_+_92499044 | 0.01 |
ENSMUST00000214239.2
|
Olfr1413
|
olfactory receptor 1413 |
| chr4_+_62501737 | 0.01 |
ENSMUST00000098031.10
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr17_-_37430949 | 0.01 |
ENSMUST00000214994.2
ENSMUST00000216341.2 |
Olfr92
|
olfactory receptor 92 |
| chr7_-_103092357 | 0.01 |
ENSMUST00000080660.6
|
Olfr605
|
olfactory receptor 605 |
| chr17_+_69746321 | 0.01 |
ENSMUST00000169935.2
|
Akain1
|
A kinase (PRKA) anchor inhibitor 1 |
| chr1_-_90771638 | 0.01 |
ENSMUST00000130846.9
|
Col6a3
|
collagen, type VI, alpha 3 |
| chr4_-_63072367 | 0.01 |
ENSMUST00000030041.5
|
Ambp
|
alpha 1 microglobulin/bikunin precursor |
| chr10_-_128974829 | 0.01 |
ENSMUST00000203887.3
ENSMUST00000204250.3 ENSMUST00000204712.4 |
Olfr770
|
olfactory receptor 770 |
| chr1_-_90771674 | 0.01 |
ENSMUST00000097653.11
ENSMUST00000188587.7 ENSMUST00000056925.16 ENSMUST00000187753.2 |
Col6a3
|
collagen, type VI, alpha 3 |
| chr11_-_99241924 | 0.01 |
ENSMUST00000017732.3
|
Krt27
|
keratin 27 |
Network of associatons between targets according to the STRING database.
First level regulatory network of En2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.1 | 1.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.2 | GO:1903632 | positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.1 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 0.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 0.8 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.2 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 2.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:1904828 | regulation of hydrogen sulfide biosynthetic process(GO:1904826) positive regulation of hydrogen sulfide biosynthetic process(GO:1904828) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0007527 | adult somatic muscle development(GO:0007527) |
| 0.0 | 0.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.1 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.0 | 0.6 | GO:0030497 | fatty acid elongation(GO:0030497) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.2 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.0 | 0.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.1 | GO:0061033 | bronchiole development(GO:0060435) bud outgrowth involved in lung branching(GO:0060447) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.6 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.3 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 0.5 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 3.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0003681 | bent DNA binding(GO:0003681) |
| 0.1 | 0.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |