Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
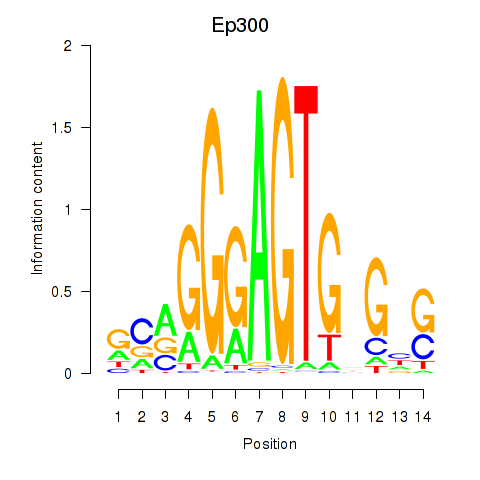
Results for Ep300
Z-value: 0.58
Transcription factors associated with Ep300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ep300
|
ENSMUSG00000055024.13 | E1A binding protein p300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ep300 | mm39_v1_chr15_+_81511486_81511626 | -0.75 | 1.5e-01 | Click! |
Activity profile of Ep300 motif
Sorted Z-values of Ep300 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_55493325 | 0.39 |
ENSMUST00000079663.7
|
Gm2174
|
predicted gene 2174 |
| chr12_-_80306865 | 0.33 |
ENSMUST00000167327.2
|
Actn1
|
actinin, alpha 1 |
| chr13_-_30168374 | 0.32 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr17_+_35780977 | 0.32 |
ENSMUST00000174525.8
ENSMUST00000068291.7 |
H2-Q10
|
histocompatibility 2, Q region locus 10 |
| chr4_+_150321142 | 0.29 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr12_-_80307110 | 0.28 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr18_-_43610829 | 0.27 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr15_+_6416229 | 0.25 |
ENSMUST00000110664.9
ENSMUST00000110663.9 ENSMUST00000161812.8 ENSMUST00000160134.8 |
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr8_+_39472981 | 0.25 |
ENSMUST00000239508.1
ENSMUST00000239509.1 |
TUSC3
|
tumor suppressor candidate 3 |
| chr11_+_53661251 | 0.24 |
ENSMUST00000138913.8
ENSMUST00000123376.8 ENSMUST00000019043.13 ENSMUST00000133291.3 |
Irf1
|
interferon regulatory factor 1 |
| chr7_+_140462343 | 0.24 |
ENSMUST00000163610.9
ENSMUST00000164681.8 |
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr4_+_150321659 | 0.24 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr10_+_79716876 | 0.23 |
ENSMUST00000166201.2
|
Prtn3
|
proteinase 3 |
| chr7_+_140461860 | 0.20 |
ENSMUST00000026560.14
|
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr11_+_53660834 | 0.20 |
ENSMUST00000108920.10
ENSMUST00000140866.9 ENSMUST00000108922.9 |
Irf1
|
interferon regulatory factor 1 |
| chr8_-_11600689 | 0.20 |
ENSMUST00000049461.7
|
Cars2
|
cysteinyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr5_-_137530214 | 0.20 |
ENSMUST00000140139.2
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr3_-_88857578 | 0.19 |
ENSMUST00000174402.8
ENSMUST00000174077.8 |
Dap3
|
death associated protein 3 |
| chr8_-_122556219 | 0.19 |
ENSMUST00000174717.8
ENSMUST00000174192.2 |
Klhdc4
|
kelch domain containing 4 |
| chr13_-_58276353 | 0.19 |
ENSMUST00000007980.7
|
Hnrnpa0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr8_-_85413707 | 0.19 |
ENSMUST00000238301.2
|
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr18_+_68433422 | 0.18 |
ENSMUST00000009679.11
ENSMUST00000131075.8 ENSMUST00000025427.14 ENSMUST00000139111.2 |
Rnmt
|
RNA (guanine-7-) methyltransferase |
| chr3_-_88858402 | 0.18 |
ENSMUST00000173021.8
|
Dap3
|
death associated protein 3 |
| chr2_-_25101354 | 0.17 |
ENSMUST00000059849.15
|
Nelfb
|
negative elongation factor complex member B |
| chr2_+_71219561 | 0.17 |
ENSMUST00000028408.3
|
Hat1
|
histone aminotransferase 1 |
| chr10_-_17898938 | 0.17 |
ENSMUST00000220110.2
|
Abracl
|
ABRA C-terminal like |
| chr19_+_18690589 | 0.17 |
ENSMUST00000055792.8
|
D030056L22Rik
|
RIKEN cDNA D030056L22 gene |
| chr4_+_149569717 | 0.17 |
ENSMUST00000030842.8
|
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr10_-_117681864 | 0.16 |
ENSMUST00000064667.9
|
Rap1b
|
RAS related protein 1b |
| chr19_+_18690556 | 0.16 |
ENSMUST00000062753.3
|
D030056L22Rik
|
RIKEN cDNA D030056L22 gene |
| chr17_-_29483075 | 0.16 |
ENSMUST00000024802.10
|
Ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr13_+_120151982 | 0.15 |
ENSMUST00000179869.3
ENSMUST00000224188.2 |
Hmgcs1
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| chr15_+_6416079 | 0.15 |
ENSMUST00000080880.12
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr10_-_57408512 | 0.15 |
ENSMUST00000169122.8
|
Serinc1
|
serine incorporator 1 |
| chr19_+_44994905 | 0.14 |
ENSMUST00000026227.3
|
Twnk
|
twinkle mtDNA helicase |
| chr8_-_85414220 | 0.14 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr11_-_113456568 | 0.14 |
ENSMUST00000071539.10
ENSMUST00000106633.10 ENSMUST00000042657.16 ENSMUST00000149034.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr17_-_24292453 | 0.14 |
ENSMUST00000017090.6
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr11_-_93859064 | 0.14 |
ENSMUST00000107844.3
ENSMUST00000170303.2 |
Nme1
Gm20390
|
NME/NM23 nucleoside diphosphate kinase 1 predicted gene 20390 |
| chr6_-_29216341 | 0.14 |
ENSMUST00000162099.8
ENSMUST00000159124.8 |
Impdh1
|
inosine monophosphate dehydrogenase 1 |
| chr19_+_44994094 | 0.14 |
ENSMUST00000236685.2
|
Twnk
|
twinkle mtDNA helicase |
| chr12_-_101943134 | 0.14 |
ENSMUST00000221227.2
|
Ndufb1
|
NADH:ubiquinone oxidoreductase subunit B1 |
| chr2_+_163444214 | 0.14 |
ENSMUST00000171696.8
ENSMUST00000109408.10 |
Ttpal
|
tocopherol (alpha) transfer protein-like |
| chr7_-_126391657 | 0.13 |
ENSMUST00000032936.8
|
Ppp4c
|
protein phosphatase 4, catalytic subunit |
| chr1_-_37469037 | 0.13 |
ENSMUST00000027286.7
|
Coa5
|
cytochrome C oxidase assembly factor 5 |
| chr8_-_48128164 | 0.13 |
ENSMUST00000080353.3
|
Ing2
|
inhibitor of growth family, member 2 |
| chr8_+_86567600 | 0.13 |
ENSMUST00000053771.14
ENSMUST00000161850.8 |
Phkb
|
phosphorylase kinase beta |
| chr7_-_140462221 | 0.13 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr6_+_134988572 | 0.13 |
ENSMUST00000032326.11
ENSMUST00000130851.8 ENSMUST00000205244.3 ENSMUST00000205055.3 ENSMUST00000204646.3 ENSMUST00000154558.3 |
Ddx47
|
DEAD box helicase 47 |
| chr4_+_118266582 | 0.13 |
ENSMUST00000144577.2
|
Med8
|
mediator complex subunit 8 |
| chr3_+_144824325 | 0.12 |
ENSMUST00000098538.9
ENSMUST00000106192.9 ENSMUST00000098539.7 ENSMUST00000029920.15 |
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr2_+_120439858 | 0.12 |
ENSMUST00000124187.8
|
Haus2
|
HAUS augmin-like complex, subunit 2 |
| chr9_+_74945101 | 0.12 |
ENSMUST00000167885.8
ENSMUST00000169188.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr9_+_20914211 | 0.12 |
ENSMUST00000214124.2
ENSMUST00000216818.2 |
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr6_+_146789978 | 0.12 |
ENSMUST00000016631.14
ENSMUST00000203730.3 ENSMUST00000111623.9 |
Ppfibp1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr11_-_84761472 | 0.11 |
ENSMUST00000018547.9
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr14_-_70396859 | 0.11 |
ENSMUST00000058240.14
ENSMUST00000153871.2 |
9930012K11Rik
|
RIKEN cDNA 9930012K11 gene |
| chr2_+_29780122 | 0.11 |
ENSMUST00000113762.8
ENSMUST00000113765.8 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chrX_-_106859842 | 0.11 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr2_-_152673585 | 0.11 |
ENSMUST00000156688.2
ENSMUST00000007803.12 |
Bcl2l1
|
BCL2-like 1 |
| chr14_+_66074751 | 0.10 |
ENSMUST00000022614.7
|
Ccdc25
|
coiled-coil domain containing 25 |
| chr15_+_8138805 | 0.10 |
ENSMUST00000230017.2
|
Nup155
|
nucleoporin 155 |
| chr12_+_78273356 | 0.10 |
ENSMUST00000110388.10
|
Gphn
|
gephyrin |
| chr14_-_32907023 | 0.10 |
ENSMUST00000130509.10
ENSMUST00000061753.15 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr10_+_63079667 | 0.10 |
ENSMUST00000020258.10
ENSMUST00000219577.2 |
Herc4
|
hect domain and RLD 4 |
| chr10_+_85665234 | 0.10 |
ENSMUST00000217667.2
|
Pwp1
|
PWP1 homolog, endonuclein |
| chr1_-_181039509 | 0.10 |
ENSMUST00000162819.9
ENSMUST00000237749.2 |
Wdr26
|
WD repeat domain 26 |
| chr16_-_94327689 | 0.10 |
ENSMUST00000023615.7
|
Vps26c
|
VPS26 endosomal protein sorting factor C |
| chr8_+_70686836 | 0.10 |
ENSMUST00000164403.8
ENSMUST00000093458.11 |
Sugp2
|
SURP and G patch domain containing 2 |
| chr8_+_61446221 | 0.10 |
ENSMUST00000120689.8
ENSMUST00000034065.14 ENSMUST00000211256.2 ENSMUST00000211672.2 |
Nek1
|
NIMA (never in mitosis gene a)-related expressed kinase 1 |
| chr19_+_8713156 | 0.10 |
ENSMUST00000210512.2
ENSMUST00000049424.11 |
Wdr74
|
WD repeat domain 74 |
| chr7_+_45219766 | 0.10 |
ENSMUST00000120864.10
|
Bcat2
|
branched chain aminotransferase 2, mitochondrial |
| chr5_+_76331727 | 0.10 |
ENSMUST00000031144.14
|
Tmem165
|
transmembrane protein 165 |
| chr4_+_86493905 | 0.10 |
ENSMUST00000091064.8
|
Rraga
|
Ras-related GTP binding A |
| chr3_-_107992662 | 0.09 |
ENSMUST00000078912.7
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr8_-_122556258 | 0.09 |
ENSMUST00000045884.17
|
Klhdc4
|
kelch domain containing 4 |
| chr4_+_149569672 | 0.09 |
ENSMUST00000124413.8
ENSMUST00000141293.8 |
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr11_-_117673008 | 0.09 |
ENSMUST00000152304.3
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr2_+_153583194 | 0.09 |
ENSMUST00000028981.9
|
Mapre1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr3_-_88857707 | 0.09 |
ENSMUST00000090938.11
|
Dap3
|
death associated protein 3 |
| chr10_-_17898838 | 0.09 |
ENSMUST00000220433.2
|
Abracl
|
ABRA C-terminal like |
| chr3_-_90340830 | 0.09 |
ENSMUST00000029542.12
|
Ints3
|
integrator complex subunit 3 |
| chr15_+_44482944 | 0.09 |
ENSMUST00000022964.9
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr11_-_102771806 | 0.09 |
ENSMUST00000107060.8
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr13_-_119545479 | 0.09 |
ENSMUST00000223268.2
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr12_-_4283926 | 0.08 |
ENSMUST00000111169.10
ENSMUST00000020981.12 |
Cenpo
|
centromere protein O |
| chr7_+_142622986 | 0.08 |
ENSMUST00000060433.10
ENSMUST00000133410.3 ENSMUST00000105920.8 ENSMUST00000177841.8 ENSMUST00000147995.2 |
Tssc4
|
tumor-suppressing subchromosomal transferable fragment 4 |
| chr19_-_41969558 | 0.08 |
ENSMUST00000026168.9
ENSMUST00000171561.8 |
Mms19
|
MMS19 cytosolic iron-sulfur assembly component |
| chr8_-_70686746 | 0.08 |
ENSMUST00000130319.2
|
Armc6
|
armadillo repeat containing 6 |
| chr2_+_29780073 | 0.08 |
ENSMUST00000028128.13
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr11_-_84761538 | 0.08 |
ENSMUST00000170741.2
ENSMUST00000172405.8 ENSMUST00000100686.10 ENSMUST00000108081.9 |
Ggnbp2
|
gametogenetin binding protein 2 |
| chr3_+_88461059 | 0.08 |
ENSMUST00000008748.8
|
Ubqln4
|
ubiquilin 4 |
| chr15_+_76215431 | 0.08 |
ENSMUST00000023221.13
|
Gpaa1
|
GPI anchor attachment protein 1 |
| chr13_+_73752125 | 0.08 |
ENSMUST00000022102.9
|
Clptm1l
|
CLPTM1-like |
| chr11_-_40646090 | 0.08 |
ENSMUST00000020576.8
|
Ccng1
|
cyclin G1 |
| chr3_-_51184730 | 0.08 |
ENSMUST00000195432.2
ENSMUST00000091144.11 ENSMUST00000156983.3 |
Elf2
|
E74-like factor 2 |
| chr11_-_120042019 | 0.08 |
ENSMUST00000179094.8
ENSMUST00000103018.11 ENSMUST00000045402.14 ENSMUST00000076697.13 ENSMUST00000053692.9 |
Slc38a10
|
solute carrier family 38, member 10 |
| chr9_-_20888054 | 0.08 |
ENSMUST00000054197.7
|
S1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr11_+_49135018 | 0.08 |
ENSMUST00000167400.8
ENSMUST00000081794.7 |
Mgat1
|
mannoside acetylglucosaminyltransferase 1 |
| chr14_+_56905698 | 0.08 |
ENSMUST00000116468.2
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr11_-_84761637 | 0.08 |
ENSMUST00000168434.8
|
Ggnbp2
|
gametogenetin binding protein 2 |
| chr18_+_24737009 | 0.07 |
ENSMUST00000234266.2
ENSMUST00000025120.8 |
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr9_-_79700789 | 0.07 |
ENSMUST00000120690.2
|
Tmem30a
|
transmembrane protein 30A |
| chr13_-_119545520 | 0.07 |
ENSMUST00000069902.13
ENSMUST00000099149.10 ENSMUST00000109204.8 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr17_+_36152559 | 0.07 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr2_+_120294046 | 0.07 |
ENSMUST00000028749.15
ENSMUST00000110721.9 ENSMUST00000239364.2 |
Capn3
|
calpain 3 |
| chr3_-_90340910 | 0.07 |
ENSMUST00000196530.2
|
Ints3
|
integrator complex subunit 3 |
| chr15_+_36174156 | 0.07 |
ENSMUST00000180159.8
ENSMUST00000057177.7 |
Polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr2_+_24276545 | 0.07 |
ENSMUST00000127242.2
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr2_+_24276616 | 0.07 |
ENSMUST00000166388.2
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr2_+_166857113 | 0.07 |
ENSMUST00000018143.16
ENSMUST00000176066.8 ENSMUST00000150571.2 |
Ddx27
|
DEAD box helicase 27 |
| chr1_+_171238911 | 0.07 |
ENSMUST00000160486.8
|
Usf1
|
upstream transcription factor 1 |
| chr8_-_106692668 | 0.07 |
ENSMUST00000116429.9
ENSMUST00000034370.17 |
Slc12a4
|
solute carrier family 12, member 4 |
| chr7_-_140462187 | 0.07 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr1_+_37469220 | 0.07 |
ENSMUST00000114925.10
ENSMUST00000027285.13 ENSMUST00000144617.8 ENSMUST00000193979.6 ENSMUST00000118059.3 ENSMUST00000193713.2 |
Unc50
|
unc-50 homolog |
| chr19_+_47056161 | 0.07 |
ENSMUST00000026027.7
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr2_+_32665781 | 0.07 |
ENSMUST00000066352.6
|
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr15_-_97665524 | 0.07 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr19_-_46327024 | 0.07 |
ENSMUST00000236046.2
ENSMUST00000236980.2 ENSMUST00000235485.2 ENSMUST00000236061.2 ENSMUST00000236236.2 ENSMUST00000236768.2 ENSMUST00000236651.2 |
Cuedc2
|
CUE domain containing 2 |
| chr9_+_74945021 | 0.06 |
ENSMUST00000168301.2
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr6_+_117894242 | 0.06 |
ENSMUST00000180020.8
ENSMUST00000177570.2 |
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr11_+_5905693 | 0.06 |
ENSMUST00000002818.9
|
Ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr3_-_129625023 | 0.06 |
ENSMUST00000029643.15
|
Gar1
|
GAR1 ribonucleoprotein |
| chr11_-_70128462 | 0.06 |
ENSMUST00000100950.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr18_-_24736848 | 0.06 |
ENSMUST00000070726.10
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr14_+_54924439 | 0.06 |
ENSMUST00000227269.2
|
1700123O20Rik
|
RIKEN cDNA 1700123O20 gene |
| chr10_-_83484467 | 0.06 |
ENSMUST00000146876.9
ENSMUST00000176294.2 |
Appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr7_+_48438751 | 0.06 |
ENSMUST00000118927.8
ENSMUST00000125280.8 |
Zdhhc13
|
zinc finger, DHHC domain containing 13 |
| chr14_-_20027112 | 0.06 |
ENSMUST00000159028.8
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr5_+_76736514 | 0.06 |
ENSMUST00000121979.8
|
Cep135
|
centrosomal protein 135 |
| chr1_-_84817000 | 0.06 |
ENSMUST00000186648.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr18_-_68433398 | 0.06 |
ENSMUST00000042852.7
|
Fam210a
|
family with sequence similarity 210, member A |
| chr2_-_152673032 | 0.06 |
ENSMUST00000128172.3
|
Bcl2l1
|
BCL2-like 1 |
| chr1_-_183078488 | 0.06 |
ENSMUST00000057062.12
|
Brox
|
BRO1 domain and CAAX motif containing |
| chr10_-_17898977 | 0.06 |
ENSMUST00000020002.9
|
Abracl
|
ABRA C-terminal like |
| chr8_-_86567506 | 0.06 |
ENSMUST00000034140.9
|
Itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr7_+_142623241 | 0.06 |
ENSMUST00000137856.2
|
Tssc4
|
tumor-suppressing subchromosomal transferable fragment 4 |
| chr3_-_63872189 | 0.05 |
ENSMUST00000029402.15
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr9_+_57847387 | 0.05 |
ENSMUST00000043059.9
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A |
| chr15_+_44482545 | 0.05 |
ENSMUST00000227691.2
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr1_+_121358778 | 0.05 |
ENSMUST00000036025.16
ENSMUST00000112621.2 |
Ccdc93
|
coiled-coil domain containing 93 |
| chr4_+_59805829 | 0.05 |
ENSMUST00000030080.7
|
Snx30
|
sorting nexin family member 30 |
| chrX_-_7834057 | 0.05 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr8_-_65302573 | 0.05 |
ENSMUST00000210166.2
|
Klhl2
|
kelch-like 2, Mayven |
| chr2_-_155571279 | 0.05 |
ENSMUST00000040833.5
|
Edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chrX_-_73289970 | 0.05 |
ENSMUST00000130007.8
|
Flna
|
filamin, alpha |
| chr7_+_65343156 | 0.05 |
ENSMUST00000032726.14
ENSMUST00000107495.5 ENSMUST00000143508.3 ENSMUST00000129166.3 ENSMUST00000206517.2 ENSMUST00000206837.2 ENSMUST00000206628.2 ENSMUST00000206361.2 |
Tm2d3
|
TM2 domain containing 3 |
| chr19_-_46327071 | 0.05 |
ENSMUST00000235977.2
ENSMUST00000167861.8 ENSMUST00000051234.9 ENSMUST00000236066.2 |
Cuedc2
|
CUE domain containing 2 |
| chr2_-_120439981 | 0.04 |
ENSMUST00000133612.2
ENSMUST00000102498.8 ENSMUST00000102499.8 |
Lrrc57
|
leucine rich repeat containing 57 |
| chr15_+_44482667 | 0.04 |
ENSMUST00000228648.2
ENSMUST00000226165.2 |
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr6_-_83483868 | 0.04 |
ENSMUST00000014698.10
ENSMUST00000113888.3 |
Dguok
|
deoxyguanosine kinase |
| chr14_+_54701594 | 0.04 |
ENSMUST00000022782.10
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr9_+_44966464 | 0.04 |
ENSMUST00000114664.8
|
Mpzl3
|
myelin protein zero-like 3 |
| chr6_+_86415342 | 0.04 |
ENSMUST00000050497.14
ENSMUST00000203568.3 |
C87436
|
expressed sequence C87436 |
| chr13_-_100867398 | 0.04 |
ENSMUST00000225990.2
ENSMUST00000091299.8 |
Cdk7
|
cyclin-dependent kinase 7 |
| chr7_+_45683122 | 0.04 |
ENSMUST00000033121.7
|
Nomo1
|
nodal modulator 1 |
| chr11_+_78136569 | 0.04 |
ENSMUST00000002133.9
|
Sdf2
|
stromal cell derived factor 2 |
| chr12_+_78273144 | 0.04 |
ENSMUST00000052472.6
|
Gphn
|
gephyrin |
| chr4_-_148123223 | 0.04 |
ENSMUST00000030879.12
ENSMUST00000137724.8 |
Clcn6
|
chloride channel, voltage-sensitive 6 |
| chr2_+_29779750 | 0.04 |
ENSMUST00000113763.8
ENSMUST00000113757.8 ENSMUST00000113756.8 ENSMUST00000133233.8 ENSMUST00000113759.9 ENSMUST00000113755.8 ENSMUST00000137558.8 ENSMUST00000046571.14 |
Odf2
|
outer dense fiber of sperm tails 2 |
| chr8_+_106877025 | 0.04 |
ENSMUST00000212963.2
ENSMUST00000034377.8 |
Pla2g15
|
phospholipase A2, group XV |
| chr2_-_102230602 | 0.04 |
ENSMUST00000152929.2
|
Trim44
|
tripartite motif-containing 44 |
| chr14_-_54647647 | 0.04 |
ENSMUST00000228488.2
ENSMUST00000195970.5 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr6_+_113508636 | 0.04 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr6_+_117893942 | 0.04 |
ENSMUST00000179478.8
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr9_-_79700660 | 0.04 |
ENSMUST00000034878.12
|
Tmem30a
|
transmembrane protein 30A |
| chr14_-_20026761 | 0.04 |
ENSMUST00000161247.2
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr3_-_63872079 | 0.04 |
ENSMUST00000161659.8
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr9_+_40103598 | 0.04 |
ENSMUST00000026693.14
ENSMUST00000168832.2 |
Zfp202
|
zinc finger protein 202 |
| chr4_+_150321272 | 0.04 |
ENSMUST00000080926.13
|
Eno1
|
enolase 1, alpha non-neuron |
| chr4_-_141602190 | 0.04 |
ENSMUST00000036854.4
|
Efhd2
|
EF hand domain containing 2 |
| chrX_-_73290140 | 0.03 |
ENSMUST00000101454.9
ENSMUST00000033699.13 |
Flna
|
filamin, alpha |
| chr18_-_24736521 | 0.03 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr16_-_52272828 | 0.03 |
ENSMUST00000170035.8
ENSMUST00000164728.8 ENSMUST00000168071.2 |
Alcam
|
activated leukocyte cell adhesion molecule |
| chr2_+_69619991 | 0.03 |
ENSMUST00000112266.8
|
Phospho2
|
phosphatase, orphan 2 |
| chr13_-_55510595 | 0.03 |
ENSMUST00000021940.8
|
Lman2
|
lectin, mannose-binding 2 |
| chr5_+_129970882 | 0.03 |
ENSMUST00000201855.2
ENSMUST00000073945.6 |
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr6_-_116605882 | 0.03 |
ENSMUST00000057540.6
|
Zfp422
|
zinc finger protein 422 |
| chr2_+_30282414 | 0.03 |
ENSMUST00000123202.8
ENSMUST00000113612.10 |
Dolpp1
|
dolichyl pyrophosphate phosphatase 1 |
| chr15_-_83054698 | 0.03 |
ENSMUST00000162178.8
|
Cyb5r3
|
cytochrome b5 reductase 3 |
| chr9_-_20864096 | 0.03 |
ENSMUST00000004202.17
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr7_-_79570342 | 0.03 |
ENSMUST00000075657.8
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr19_-_44994824 | 0.02 |
ENSMUST00000097715.4
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr11_-_93846453 | 0.02 |
ENSMUST00000072566.5
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr2_+_69619967 | 0.02 |
ENSMUST00000151298.8
ENSMUST00000028494.9 |
Phospho2
|
phosphatase, orphan 2 |
| chr4_+_114914880 | 0.02 |
ENSMUST00000161601.8
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr11_+_69886652 | 0.02 |
ENSMUST00000101526.9
|
Phf23
|
PHD finger protein 23 |
| chr11_-_70128587 | 0.02 |
ENSMUST00000108576.10
|
0610010K14Rik
|
RIKEN cDNA 0610010K14 gene |
| chr10_-_127047396 | 0.02 |
ENSMUST00000013970.9
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr16_-_13804699 | 0.02 |
ENSMUST00000117803.2
|
Ifitm7
|
interferon induced transmembrane protein 7 |
| chr4_+_148123490 | 0.02 |
ENSMUST00000097788.11
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr2_+_29779721 | 0.02 |
ENSMUST00000113767.8
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr15_-_97665679 | 0.02 |
ENSMUST00000129223.9
ENSMUST00000126854.9 ENSMUST00000135080.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr4_+_3938903 | 0.02 |
ENSMUST00000121210.8
ENSMUST00000121651.8 ENSMUST00000041122.11 ENSMUST00000120732.8 ENSMUST00000119307.8 ENSMUST00000123769.8 |
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr4_-_3938352 | 0.02 |
ENSMUST00000003369.10
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chrX_+_149830166 | 0.02 |
ENSMUST00000026296.8
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr6_-_113508536 | 0.02 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr2_-_120439826 | 0.02 |
ENSMUST00000102497.10
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr6_+_125048230 | 0.02 |
ENSMUST00000140346.9
ENSMUST00000171989.3 |
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr1_+_74811045 | 0.02 |
ENSMUST00000006716.8
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ep300
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035740 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.4 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.2 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.3 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.0 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.0 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |