Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
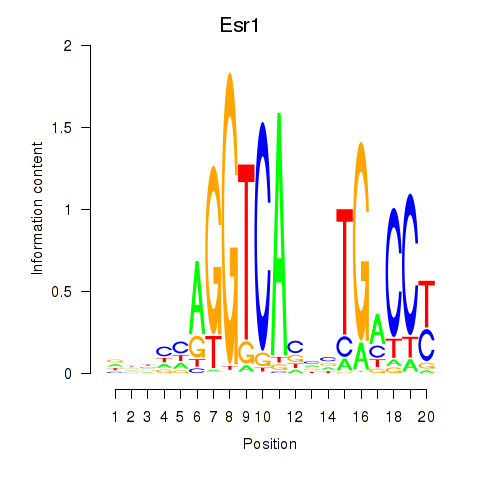
Results for Esr1
Z-value: 0.67
Transcription factors associated with Esr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esr1
|
ENSMUSG00000019768.17 | estrogen receptor 1 (alpha) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esr1 | mm39_v1_chr10_+_4561974_4562075 | 0.97 | 6.4e-03 | Click! |
Activity profile of Esr1 motif
Sorted Z-values of Esr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_110848339 | 0.73 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr11_-_102255999 | 0.34 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr16_+_22926162 | 0.28 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr7_+_125202653 | 0.26 |
ENSMUST00000206103.2
ENSMUST00000033000.8 |
Il21r
|
interleukin 21 receptor |
| chr7_-_144493560 | 0.25 |
ENSMUST00000093962.5
|
Ccnd1
|
cyclin D1 |
| chr2_-_50186690 | 0.23 |
ENSMUST00000144143.8
ENSMUST00000102769.11 ENSMUST00000133768.2 |
Mmadhc
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr11_+_87684299 | 0.22 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr17_+_28075415 | 0.20 |
ENSMUST00000114849.3
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1 |
| chr11_+_87684548 | 0.19 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr19_+_3372296 | 0.19 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr11_+_23615612 | 0.18 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr6_-_69415741 | 0.18 |
ENSMUST00000103354.3
|
Igkv4-59
|
immunoglobulin kappa variable 4-59 |
| chr6_-_69584812 | 0.18 |
ENSMUST00000103359.3
|
Igkv4-55
|
immunoglobulin kappa variable 4-55 |
| chr5_+_3621673 | 0.17 |
ENSMUST00000008451.12
|
1700109H08Rik
|
RIKEN cDNA 1700109H08 gene |
| chr5_-_113428407 | 0.17 |
ENSMUST00000112324.2
ENSMUST00000057209.12 |
Sgsm1
|
small G protein signaling modulator 1 |
| chr8_+_72021567 | 0.16 |
ENSMUST00000034267.5
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr1_-_63215812 | 0.16 |
ENSMUST00000185847.2
ENSMUST00000185732.7 ENSMUST00000188370.7 ENSMUST00000168099.9 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr5_+_122347912 | 0.15 |
ENSMUST00000143560.8
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr7_-_140462187 | 0.15 |
ENSMUST00000211179.2
|
Sirt3
|
sirtuin 3 |
| chr8_+_72021510 | 0.15 |
ENSMUST00000212889.2
|
Slc27a1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr11_+_87685032 | 0.15 |
ENSMUST00000121303.8
|
Mpo
|
myeloperoxidase |
| chr7_+_46496929 | 0.15 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr14_-_56322654 | 0.14 |
ENSMUST00000015594.9
|
Mcpt8
|
mast cell protease 8 |
| chr7_+_46496552 | 0.14 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr11_-_31621863 | 0.14 |
ENSMUST00000058060.14
|
Bod1
|
biorientation of chromosomes in cell division 1 |
| chr10_+_79722081 | 0.13 |
ENSMUST00000046091.7
|
Elane
|
elastase, neutrophil expressed |
| chr19_-_4240984 | 0.13 |
ENSMUST00000045864.4
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr7_-_125976580 | 0.13 |
ENSMUST00000119846.8
ENSMUST00000119754.8 ENSMUST00000032994.15 |
Spns1
|
spinster homolog 1 |
| chr9_+_107926502 | 0.13 |
ENSMUST00000047947.9
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr11_-_117764258 | 0.13 |
ENSMUST00000033230.8
|
Tha1
|
threonine aldolase 1 |
| chr7_+_144468837 | 0.13 |
ENSMUST00000128057.8
ENSMUST00000033388.12 ENSMUST00000141737.2 ENSMUST00000105895.2 |
LTO1
|
ABCE maturation factor |
| chr11_-_99045894 | 0.13 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr17_+_28491085 | 0.13 |
ENSMUST00000169040.3
|
Ppard
|
peroxisome proliferator activator receptor delta |
| chr3_+_96088467 | 0.13 |
ENSMUST00000035371.9
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr8_+_121395047 | 0.13 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr5_+_36050663 | 0.12 |
ENSMUST00000064571.11
|
Afap1
|
actin filament associated protein 1 |
| chr17_+_35539505 | 0.12 |
ENSMUST00000105041.10
ENSMUST00000073208.6 |
H2-Q1
|
histocompatibility 2, Q region locus 1 |
| chr13_-_107073415 | 0.12 |
ENSMUST00000080856.14
|
Ipo11
|
importin 11 |
| chr15_+_79784543 | 0.12 |
ENSMUST00000230741.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr4_+_99812912 | 0.12 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr4_+_140427799 | 0.12 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr10_+_39245746 | 0.12 |
ENSMUST00000063091.13
ENSMUST00000099967.10 ENSMUST00000126486.8 |
Fyn
|
Fyn proto-oncogene |
| chr10_+_80692948 | 0.12 |
ENSMUST00000220091.2
|
Sppl2b
|
signal peptide peptidase like 2B |
| chr19_-_43512929 | 0.12 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr15_+_79784365 | 0.12 |
ENSMUST00000230135.2
|
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr7_-_140462221 | 0.12 |
ENSMUST00000026559.14
|
Sirt3
|
sirtuin 3 |
| chr1_-_63215952 | 0.12 |
ENSMUST00000185412.7
ENSMUST00000027111.15 ENSMUST00000189664.2 |
Ndufs1
|
NADH:ubiquinone oxidoreductase core subunit S1 |
| chr17_+_37581103 | 0.12 |
ENSMUST00000038580.7
|
H2-M3
|
histocompatibility 2, M region locus 3 |
| chr17_-_26063488 | 0.11 |
ENSMUST00000176709.2
|
Rhot2
|
ras homolog family member T2 |
| chr14_-_30909082 | 0.11 |
ENSMUST00000170253.8
|
Nisch
|
nischarin |
| chr8_+_73488496 | 0.11 |
ENSMUST00000058099.9
|
F2rl3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr15_+_76227695 | 0.11 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr18_-_34757653 | 0.11 |
ENSMUST00000003876.10
ENSMUST00000115766.8 ENSMUST00000097626.10 ENSMUST00000115765.2 |
Brd8
|
bromodomain containing 8 |
| chr7_+_141055350 | 0.11 |
ENSMUST00000138092.8
ENSMUST00000146305.8 |
Tspan4
|
tetraspanin 4 |
| chr7_+_46496506 | 0.11 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr2_+_30331839 | 0.11 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr4_+_107659361 | 0.11 |
ENSMUST00000106731.4
|
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr7_+_140462343 | 0.11 |
ENSMUST00000163610.9
ENSMUST00000164681.8 |
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr2_+_181506130 | 0.11 |
ENSMUST00000039551.9
|
Polr3k
|
polymerase (RNA) III (DNA directed) polypeptide K |
| chr4_+_150321142 | 0.11 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr3_+_90011435 | 0.11 |
ENSMUST00000029548.9
ENSMUST00000200410.2 |
Nup210l
|
nucleoporin 210-like |
| chr10_-_127147609 | 0.11 |
ENSMUST00000037290.12
ENSMUST00000171564.8 |
Mars1
|
methionine-tRNA synthetase 1 |
| chr11_-_116164928 | 0.11 |
ENSMUST00000106425.4
|
Srp68
|
signal recognition particle 68 |
| chr3_-_19319155 | 0.10 |
ENSMUST00000091314.11
|
Pde7a
|
phosphodiesterase 7A |
| chr9_+_37119472 | 0.10 |
ENSMUST00000034632.10
|
Tmem218
|
transmembrane protein 218 |
| chr4_-_150993886 | 0.10 |
ENSMUST00000128075.8
|
Park7
|
Parkinson disease (autosomal recessive, early onset) 7 |
| chr11_+_69737200 | 0.10 |
ENSMUST00000108632.8
|
Plscr3
|
phospholipid scramblase 3 |
| chr3_-_59118293 | 0.10 |
ENSMUST00000040622.3
|
P2ry13
|
purinergic receptor P2Y, G-protein coupled 13 |
| chr5_+_137517140 | 0.10 |
ENSMUST00000031727.10
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr16_+_16120810 | 0.10 |
ENSMUST00000159962.8
ENSMUST00000059955.15 |
Yars2
|
tyrosyl-tRNA synthetase 2 (mitochondrial) |
| chr10_-_81201642 | 0.10 |
ENSMUST00000020456.5
|
4930404N11Rik
|
RIKEN cDNA 4930404N11 gene |
| chr11_-_59397352 | 0.10 |
ENSMUST00000013262.15
ENSMUST00000101150.9 |
Zkscan17
|
zinc finger with KRAB and SCAN domains 17 |
| chr16_-_20245138 | 0.10 |
ENSMUST00000079158.13
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr18_-_38130581 | 0.10 |
ENSMUST00000042944.9
|
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chrX_+_73468140 | 0.10 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr19_+_4806544 | 0.10 |
ENSMUST00000182821.8
ENSMUST00000036744.8 |
Rbm4b
|
RNA binding motif protein 4B |
| chr9_-_55419442 | 0.10 |
ENSMUST00000034866.9
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide |
| chr3_-_95125190 | 0.10 |
ENSMUST00000136139.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr15_-_77527470 | 0.10 |
ENSMUST00000181154.2
ENSMUST00000180949.8 ENSMUST00000166623.10 |
Apol11b
|
apolipoprotein L 11b |
| chr4_-_59783780 | 0.10 |
ENSMUST00000107526.8
ENSMUST00000095063.11 |
Inip
|
INTS3 and NABP interacting protein |
| chr2_-_101451383 | 0.09 |
ENSMUST00000090513.11
|
Iftap
|
intraflagellar transport associated protein |
| chr14_+_66534478 | 0.09 |
ENSMUST00000022623.13
|
Trim35
|
tripartite motif-containing 35 |
| chr9_-_64160899 | 0.09 |
ENSMUST00000005066.9
|
Map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr7_+_28416270 | 0.09 |
ENSMUST00000108279.9
|
Fbxo17
|
F-box protein 17 |
| chr2_-_26270612 | 0.09 |
ENSMUST00000114115.9
ENSMUST00000035427.11 |
Snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr7_+_28416194 | 0.09 |
ENSMUST00000032818.13
|
Fbxo17
|
F-box protein 17 |
| chr1_-_160898181 | 0.09 |
ENSMUST00000035430.4
|
Dars2
|
aspartyl-tRNA synthetase 2 (mitochondrial) |
| chr4_-_132072988 | 0.09 |
ENSMUST00000030726.13
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr19_-_5776268 | 0.09 |
ENSMUST00000075606.6
ENSMUST00000236215.2 ENSMUST00000235730.2 ENSMUST00000237081.2 ENSMUST00000049295.15 |
Ehbp1l1
|
EH domain binding protein 1-like 1 |
| chr8_-_106660470 | 0.09 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr9_+_44308149 | 0.09 |
ENSMUST00000215420.2
|
Slc37a4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr1_+_91468409 | 0.09 |
ENSMUST00000027538.9
ENSMUST00000190484.7 ENSMUST00000186068.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr12_+_55431007 | 0.09 |
ENSMUST00000163070.8
|
Psma6
|
proteasome subunit alpha 6 |
| chr7_-_143376529 | 0.09 |
ENSMUST00000156779.2
ENSMUST00000033415.15 |
Nadsyn1
|
NAD synthetase 1 |
| chr15_-_102097387 | 0.09 |
ENSMUST00000230288.2
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr12_-_69771604 | 0.08 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr8_-_106015682 | 0.08 |
ENSMUST00000212922.2
ENSMUST00000212219.2 |
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene |
| chr6_-_70383976 | 0.08 |
ENSMUST00000103393.2
|
Igkv6-15
|
immunoglobulin kappa variable 6-15 |
| chr3_+_88744323 | 0.08 |
ENSMUST00000081695.14
ENSMUST00000090942.6 |
Gon4l
|
gon-4-like (C.elegans) |
| chr4_+_49521176 | 0.08 |
ENSMUST00000042964.13
ENSMUST00000107696.2 |
Zfp189
|
zinc finger protein 189 |
| chr9_-_44253630 | 0.08 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr18_-_12254562 | 0.08 |
ENSMUST00000055447.14
ENSMUST00000209859.2 |
Tmem241
|
transmembrane protein 241 |
| chr17_-_33007238 | 0.08 |
ENSMUST00000159086.10
|
Zfp871
|
zinc finger protein 871 |
| chr4_-_132073048 | 0.08 |
ENSMUST00000084250.11
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr9_+_65583852 | 0.08 |
ENSMUST00000136166.3
|
Oaz2
|
ornithine decarboxylase antizyme 2 |
| chr10_+_81012465 | 0.08 |
ENSMUST00000047864.11
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr6_+_48653047 | 0.08 |
ENSMUST00000054050.5
|
Gimap9
|
GTPase, IMAP family member 9 |
| chr1_-_171050004 | 0.08 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr17_-_28128180 | 0.08 |
ENSMUST00000114848.8
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr1_+_127796508 | 0.08 |
ENSMUST00000037649.6
ENSMUST00000212506.2 |
Rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr10_+_62897353 | 0.08 |
ENSMUST00000178684.3
ENSMUST00000020266.15 |
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chrX_+_73348832 | 0.08 |
ENSMUST00000153141.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr6_+_87890906 | 0.08 |
ENSMUST00000032141.14
|
Hmces
|
5-hydroxymethylcytosine (hmC) binding, ES cell specific |
| chr4_-_86530245 | 0.08 |
ENSMUST00000125481.3
ENSMUST00000070607.9 |
Haus6
|
HAUS augmin-like complex, subunit 6 |
| chr6_-_128414616 | 0.08 |
ENSMUST00000151796.3
|
Fkbp4
|
FK506 binding protein 4 |
| chr6_+_116185077 | 0.08 |
ENSMUST00000204051.2
|
Washc2
|
WASH complex subunit 2` |
| chr16_-_91415873 | 0.08 |
ENSMUST00000143058.2
ENSMUST00000049244.10 ENSMUST00000169982.2 ENSMUST00000133731.2 |
Dnajc28
|
DnaJ heat shock protein family (Hsp40) member C28 |
| chr6_-_70194405 | 0.08 |
ENSMUST00000103384.2
|
Igkv8-24
|
immunoglobulin kappa chain variable 8-24 |
| chr19_-_4241034 | 0.08 |
ENSMUST00000237495.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr9_+_107926441 | 0.08 |
ENSMUST00000112295.9
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr4_+_106591080 | 0.08 |
ENSMUST00000047620.3
|
Fam151a
|
family with sequence simliarity 151, member A |
| chr7_+_140461860 | 0.08 |
ENSMUST00000026560.14
|
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr17_-_26160872 | 0.08 |
ENSMUST00000139226.2
ENSMUST00000097368.10 ENSMUST00000140304.2 ENSMUST00000026823.16 |
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_-_96993169 | 0.08 |
ENSMUST00000085206.11
|
Slc25a36
|
solute carrier family 25, member 36 |
| chrX_-_92675719 | 0.07 |
ENSMUST00000006856.3
|
Pola1
|
polymerase (DNA directed), alpha 1 |
| chr2_-_180928867 | 0.07 |
ENSMUST00000130475.8
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr11_-_78427061 | 0.07 |
ENSMUST00000017759.9
ENSMUST00000108277.3 |
Tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr3_-_95125002 | 0.07 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr6_+_116528102 | 0.07 |
ENSMUST00000122096.3
|
Eif4a3l2
|
eukaryotic translation initiation factor 4A3 like 2 |
| chrX_-_149879501 | 0.07 |
ENSMUST00000112683.9
ENSMUST00000026295.10 |
Tsr2
|
TSR2 20S rRNA accumulation |
| chr15_-_102097466 | 0.07 |
ENSMUST00000023805.3
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr2_+_130969200 | 0.07 |
ENSMUST00000099349.10
ENSMUST00000100763.9 |
Hspa12b
|
heat shock protein 12B |
| chr7_-_98790275 | 0.07 |
ENSMUST00000037968.10
|
Uvrag
|
UV radiation resistance associated gene |
| chr16_+_32002750 | 0.07 |
ENSMUST00000231512.2
|
Bex6
|
brain expressed family member 6 |
| chr9_+_21095399 | 0.07 |
ENSMUST00000115458.9
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr8_+_124170027 | 0.07 |
ENSMUST00000108830.2
|
Def8
|
differentially expressed in FDCP 8 |
| chr14_+_75253453 | 0.07 |
ENSMUST00000036072.8
|
Rubcnl
|
RUN and cysteine rich domain containing beclin 1 interacting protein like |
| chr17_-_26138480 | 0.07 |
ENSMUST00000164982.8
ENSMUST00000179998.8 ENSMUST00000167626.8 ENSMUST00000164738.2 ENSMUST00000026826.14 ENSMUST00000167018.8 |
Rab40c
|
Rab40C, member RAS oncogene family |
| chr17_-_26063391 | 0.07 |
ENSMUST00000176591.8
|
Rhot2
|
ras homolog family member T2 |
| chr2_-_164675357 | 0.07 |
ENSMUST00000042775.5
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr4_+_116565898 | 0.07 |
ENSMUST00000135499.8
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr12_+_84408803 | 0.07 |
ENSMUST00000110278.8
ENSMUST00000145522.2 |
Coq6
|
coenzyme Q6 monooxygenase |
| chr1_-_93729562 | 0.07 |
ENSMUST00000112890.3
|
Dtymk
|
deoxythymidylate kinase |
| chr11_-_70873773 | 0.07 |
ENSMUST00000078528.7
|
C1qbp
|
complement component 1, q subcomponent binding protein |
| chr17_+_25517363 | 0.07 |
ENSMUST00000037453.4
|
Prss34
|
protease, serine 34 |
| chr8_+_85415935 | 0.07 |
ENSMUST00000125370.10
ENSMUST00000175784.2 |
Trmt1
|
tRNA methyltransferase 1 |
| chr13_+_49495025 | 0.07 |
ENSMUST00000048544.14
ENSMUST00000110084.4 ENSMUST00000110085.11 |
Bicd2
|
BICD cargo adaptor 2 |
| chr6_+_3498382 | 0.07 |
ENSMUST00000001412.17
ENSMUST00000170873.10 ENSMUST00000164052.5 |
Vps50
|
VPS50 EARP/GARPII complex subunit |
| chr2_+_162773440 | 0.07 |
ENSMUST00000130411.7
ENSMUST00000126163.3 |
Srsf6
|
serine and arginine-rich splicing factor 6 |
| chr17_-_23990512 | 0.07 |
ENSMUST00000226460.2
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr19_-_46033353 | 0.07 |
ENSMUST00000026252.14
ENSMUST00000156585.9 ENSMUST00000185355.7 ENSMUST00000152946.8 |
Ldb1
|
LIM domain binding 1 |
| chr6_-_6217126 | 0.07 |
ENSMUST00000188414.4
|
Slc25a13
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 |
| chr19_+_56536685 | 0.07 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr5_+_134212836 | 0.07 |
ENSMUST00000016086.10
|
Gtf2ird2
|
GTF2I repeat domain containing 2 |
| chr17_-_28705055 | 0.07 |
ENSMUST00000233870.2
|
Fkbp5
|
FK506 binding protein 5 |
| chr7_-_126074256 | 0.07 |
ENSMUST00000205440.2
ENSMUST00000032978.8 ENSMUST00000205340.2 |
Sh2b1
|
SH2B adaptor protein 1 |
| chr16_+_22926504 | 0.07 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr2_+_163503415 | 0.07 |
ENSMUST00000135537.8
|
Pkig
|
protein kinase inhibitor, gamma |
| chr5_-_100867520 | 0.07 |
ENSMUST00000112908.2
ENSMUST00000045617.15 |
Hpse
|
heparanase |
| chr13_+_38223023 | 0.07 |
ENSMUST00000226110.2
|
Riok1
|
RIO kinase 1 |
| chr9_+_113760002 | 0.07 |
ENSMUST00000084885.12
ENSMUST00000009885.14 |
Ubp1
|
upstream binding protein 1 |
| chr9_-_42383494 | 0.07 |
ENSMUST00000128959.8
ENSMUST00000066148.12 ENSMUST00000138506.8 |
Tbcel
|
tubulin folding cofactor E-like |
| chr7_+_89814713 | 0.07 |
ENSMUST00000207084.2
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_-_121120052 | 0.07 |
ENSMUST00000169393.8
ENSMUST00000106115.8 ENSMUST00000038709.14 ENSMUST00000147490.6 |
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr14_+_30547541 | 0.07 |
ENSMUST00000006701.8
|
Stimate
|
STIM activating enhancer |
| chr4_+_21879660 | 0.07 |
ENSMUST00000029909.3
|
Coq3
|
coenzyme Q3 methyltransferase |
| chr5_+_122348140 | 0.07 |
ENSMUST00000196187.5
ENSMUST00000100747.3 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr16_+_35590745 | 0.07 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr12_-_115766700 | 0.07 |
ENSMUST00000196587.5
ENSMUST00000103543.3 |
Ighv1-74
|
immunoglobulin heavy variable V1-74 |
| chr14_-_104760051 | 0.07 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr14_-_60488922 | 0.07 |
ENSMUST00000225311.2
ENSMUST00000041905.8 |
Gm49336
|
predicted gene, 49336 |
| chr3_-_79535966 | 0.06 |
ENSMUST00000120992.8
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase |
| chrX_+_158771429 | 0.06 |
ENSMUST00000033665.9
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr2_+_28531239 | 0.06 |
ENSMUST00000028155.12
ENSMUST00000113869.8 ENSMUST00000113867.9 |
Tsc1
|
TSC complex subunit 1 |
| chr17_+_35481702 | 0.06 |
ENSMUST00000172785.8
|
H2-D1
|
histocompatibility 2, D region locus 1 |
| chr8_-_33820578 | 0.06 |
ENSMUST00000211498.2
|
Wrn
|
Werner syndrome RecQ like helicase |
| chr2_-_155668567 | 0.06 |
ENSMUST00000109638.2
ENSMUST00000134278.2 |
Eif6
|
eukaryotic translation initiation factor 6 |
| chr19_+_46316617 | 0.06 |
ENSMUST00000026256.9
ENSMUST00000177667.2 |
Fbxl15
|
F-box and leucine-rich repeat protein 15 |
| chrX_+_73352694 | 0.06 |
ENSMUST00000130581.2
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1 |
| chr9_-_119813724 | 0.06 |
ENSMUST00000213936.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr11_+_50101717 | 0.06 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr12_-_113324852 | 0.06 |
ENSMUST00000223179.2
ENSMUST00000103423.3 |
Ighg3
|
Immunoglobulin heavy constant gamma 3 |
| chr11_+_101339233 | 0.06 |
ENSMUST00000010502.13
|
Ifi35
|
interferon-induced protein 35 |
| chr2_+_22785534 | 0.06 |
ENSMUST00000053729.14
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr5_+_138115165 | 0.06 |
ENSMUST00000062350.15
ENSMUST00000110961.9 ENSMUST00000080732.10 ENSMUST00000110960.9 ENSMUST00000142185.8 ENSMUST00000136425.2 ENSMUST00000110959.2 |
Zscan21
|
zinc finger and SCAN domain containing 21 |
| chr15_+_100251030 | 0.06 |
ENSMUST00000075675.7
ENSMUST00000088142.6 ENSMUST00000176287.2 |
Methig1
Mettl7a2
|
methyltransferase hypoxia inducible domain containing 1 methyltransferase like 7A2 |
| chr7_-_30443106 | 0.06 |
ENSMUST00000182634.8
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr11_-_69553390 | 0.06 |
ENSMUST00000129224.8
ENSMUST00000155200.8 |
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr8_+_11608412 | 0.06 |
ENSMUST00000209565.2
|
Ing1
|
inhibitor of growth family, member 1 |
| chr15_+_82012114 | 0.06 |
ENSMUST00000089174.6
|
Ccdc134
|
coiled-coil domain containing 134 |
| chr18_-_56705960 | 0.06 |
ENSMUST00000174518.8
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr2_-_105734829 | 0.06 |
ENSMUST00000122965.8
|
Elp4
|
elongator acetyltransferase complex subunit 4 |
| chr8_-_112026854 | 0.06 |
ENSMUST00000038739.5
|
Rfwd3
|
ring finger and WD repeat domain 3 |
| chr10_+_79650496 | 0.06 |
ENSMUST00000218857.2
ENSMUST00000220365.2 |
Palm
|
paralemmin |
| chr14_-_52252003 | 0.06 |
ENSMUST00000226522.2
|
Zfp219
|
zinc finger protein 219 |
| chr4_-_115910577 | 0.06 |
ENSMUST00000165493.8
|
Nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr1_+_131616407 | 0.06 |
ENSMUST00000064679.9
ENSMUST00000064664.10 ENSMUST00000136247.8 |
Rab7b
|
RAB7B, member RAS oncogene family |
| chr1_-_75156993 | 0.06 |
ENSMUST00000027396.15
|
Abcb6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr6_+_70192384 | 0.06 |
ENSMUST00000103383.3
|
Igkv6-25
|
immunoglobulin kappa chain variable 6-25 |
| chr4_-_49521036 | 0.06 |
ENSMUST00000057829.4
|
Mrpl50
|
mitochondrial ribosomal protein L50 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.2 | 0.7 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 0.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0097021 | regulation of tolerance induction to self antigen(GO:0002649) lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.4 | GO:0019661 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.1 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.0 | 0.3 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0006233 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.0 | 0.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.1 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.1 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 | 0.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0015755 | fructose transport(GO:0015755) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.0 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.0 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.0 | 0.0 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.0 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.2 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.0 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.4 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 0.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.0 | 0.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.1 | GO:0015152 | glucose-6-phosphate transmembrane transporter activity(GO:0015152) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.1 | GO:0070336 | forked DNA-dependent helicase activity(GO:0061749) flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.3 | GO:0031957 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) uniporter activity(GO:0015292) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.0 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.0 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.0 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |