Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
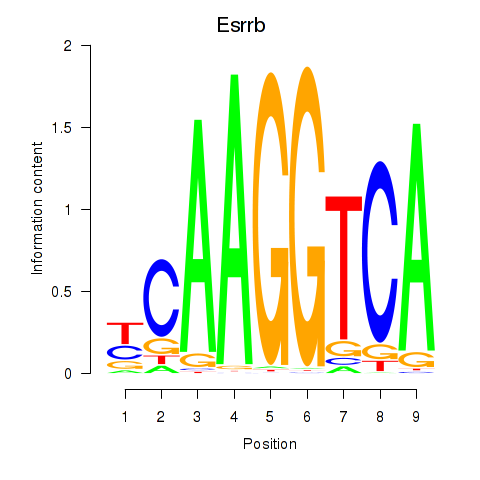
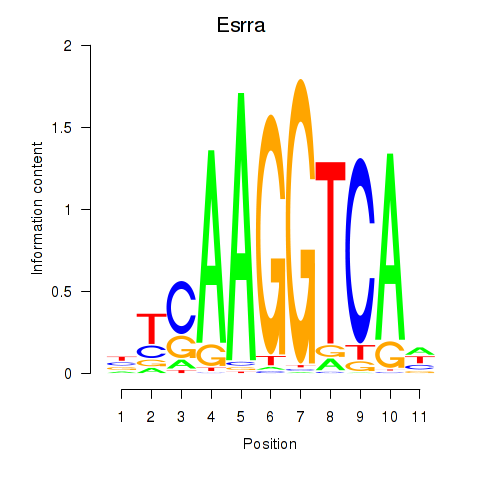
Results for Esrrb_Esrra
Z-value: 3.98
Transcription factors associated with Esrrb_Esrra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrb
|
ENSMUSG00000021255.18 | estrogen related receptor, beta |
|
Esrra
|
ENSMUSG00000024955.16 | estrogen related receptor, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrrb | mm39_v1_chr12_+_86468401_86468436 | -0.97 | 6.0e-03 | Click! |
| Esrra | mm39_v1_chr19_-_6899121_6899171 | 0.87 | 5.7e-02 | Click! |
Activity profile of Esrrb_Esrra motif
Sorted Z-values of Esrrb_Esrra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_73741664 | 5.92 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr16_+_32427738 | 4.88 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr10_+_127919142 | 4.54 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr15_+_76227695 | 4.46 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr8_+_121395047 | 3.71 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr1_-_135241429 | 3.48 |
ENSMUST00000134088.3
ENSMUST00000081104.10 |
Timm17a
|
translocase of inner mitochondrial membrane 17a |
| chr15_-_79976016 | 3.31 |
ENSMUST00000185306.3
|
Rpl3
|
ribosomal protein L3 |
| chr16_+_32427789 | 3.05 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr4_-_40279382 | 3.01 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr19_-_43512929 | 2.94 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr17_-_26087696 | 2.84 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr13_-_74498320 | 2.84 |
ENSMUST00000221594.2
ENSMUST00000022062.8 |
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chrX_+_74425990 | 2.81 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr10_-_126866682 | 2.79 |
ENSMUST00000040560.11
|
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr3_+_32791139 | 2.77 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr16_-_91728162 | 2.75 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr6_+_90527762 | 2.58 |
ENSMUST00000130418.8
ENSMUST00000032175.11 ENSMUST00000203111.2 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr5_-_115257336 | 2.45 |
ENSMUST00000031524.11
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain |
| chr7_+_46496929 | 2.44 |
ENSMUST00000132157.2
ENSMUST00000210631.2 |
Ldha
|
lactate dehydrogenase A |
| chr4_+_150321142 | 2.43 |
ENSMUST00000150175.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr7_+_120234399 | 2.34 |
ENSMUST00000033176.7
ENSMUST00000208400.2 |
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr9_+_107957640 | 2.32 |
ENSMUST00000162886.2
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr7_+_46496552 | 2.32 |
ENSMUST00000005051.6
|
Ldha
|
lactate dehydrogenase A |
| chr4_+_99812912 | 2.31 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr7_+_46496506 | 2.21 |
ENSMUST00000209984.2
|
Ldha
|
lactate dehydrogenase A |
| chr9_+_107957621 | 2.20 |
ENSMUST00000035211.14
|
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr1_-_75196496 | 2.20 |
ENSMUST00000186758.7
|
Tuba4a
|
tubulin, alpha 4A |
| chr8_+_85696695 | 2.16 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr8_+_85696453 | 2.16 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr16_-_3895642 | 2.09 |
ENSMUST00000006137.9
|
Trap1
|
TNF receptor-associated protein 1 |
| chr4_-_129436465 | 2.08 |
ENSMUST00000102597.5
|
Hdac1
|
histone deacetylase 1 |
| chr4_+_128887017 | 2.08 |
ENSMUST00000030583.13
ENSMUST00000102604.11 |
Ak2
|
adenylate kinase 2 |
| chr19_+_6449776 | 2.06 |
ENSMUST00000113468.8
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr2_-_157408239 | 2.04 |
ENSMUST00000109528.9
ENSMUST00000088494.3 |
Blcap
|
bladder cancer associated protein |
| chr15_+_44291470 | 2.03 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr8_+_121394961 | 2.00 |
ENSMUST00000034276.13
ENSMUST00000181586.8 |
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chr1_-_52230062 | 1.97 |
ENSMUST00000156887.8
ENSMUST00000129107.2 |
Gls
|
glutaminase |
| chr10_+_78410803 | 1.96 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr11_-_4045343 | 1.95 |
ENSMUST00000004868.6
|
Mtfp1
|
mitochondrial fission process 1 |
| chr8_-_95564881 | 1.94 |
ENSMUST00000034233.15
ENSMUST00000162538.9 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr13_+_54346116 | 1.93 |
ENSMUST00000038101.4
|
Hrh2
|
histamine receptor H2 |
| chr19_+_6449887 | 1.92 |
ENSMUST00000146601.8
ENSMUST00000150713.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr19_+_6450553 | 1.89 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr4_+_134123631 | 1.88 |
ENSMUST00000105869.9
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr9_-_50515089 | 1.84 |
ENSMUST00000000175.6
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr11_-_96720309 | 1.82 |
ENSMUST00000167149.8
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1 |
| chr10_-_90959817 | 1.79 |
ENSMUST00000164505.2
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr6_-_76474767 | 1.79 |
ENSMUST00000097218.7
|
Gm9008
|
predicted pseudogene 9008 |
| chr15_-_44291699 | 1.78 |
ENSMUST00000038719.8
|
Nudcd1
|
NudC domain containing 1 |
| chr8_+_85696396 | 1.78 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr9_-_121686601 | 1.76 |
ENSMUST00000213124.2
ENSMUST00000215300.2 ENSMUST00000213147.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr2_+_32477069 | 1.73 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chrX_-_47602395 | 1.72 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr12_-_71183371 | 1.72 |
ENSMUST00000221367.2
ENSMUST00000220482.2 ENSMUST00000221892.2 ENSMUST00000221178.2 ENSMUST00000221559.2 ENSMUST00000166120.9 ENSMUST00000021486.10 ENSMUST00000221797.2 ENSMUST00000221815.2 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr17_+_29251602 | 1.66 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr2_+_140012560 | 1.65 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr16_+_22926162 | 1.61 |
ENSMUST00000023599.13
ENSMUST00000168891.8 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr7_-_100232276 | 1.60 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr2_+_131028861 | 1.59 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr14_-_52252003 | 1.58 |
ENSMUST00000226522.2
|
Zfp219
|
zinc finger protein 219 |
| chr8_-_85696369 | 1.57 |
ENSMUST00000109736.9
ENSMUST00000140561.8 |
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr12_-_75678092 | 1.56 |
ENSMUST00000238938.2
|
Rplp2-ps1
|
ribosomal protein, large P2, pseudogene 1 |
| chr9_+_54493784 | 1.55 |
ENSMUST00000167866.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr10_-_71180763 | 1.55 |
ENSMUST00000045887.9
|
Cisd1
|
CDGSH iron sulfur domain 1 |
| chr7_+_28524627 | 1.54 |
ENSMUST00000066264.13
|
Ech1
|
enoyl coenzyme A hydratase 1, peroxisomal |
| chr15_-_35938155 | 1.52 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr15_+_100659622 | 1.51 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr4_-_45108038 | 1.50 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr2_-_11782381 | 1.50 |
ENSMUST00000071564.14
|
Fbh1
|
F-box DNA helicase 1 |
| chr9_-_106768601 | 1.50 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr15_-_102425241 | 1.48 |
ENSMUST00000169162.8
ENSMUST00000023812.10 ENSMUST00000165174.8 ENSMUST00000169367.8 ENSMUST00000169377.8 |
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr5_+_33493529 | 1.48 |
ENSMUST00000202113.2
|
Maea
|
macrophage erythroblast attacher |
| chr15_+_81756671 | 1.46 |
ENSMUST00000135198.2
ENSMUST00000157003.8 ENSMUST00000229068.2 |
Aco2
|
aconitase 2, mitochondrial |
| chr18_+_77861656 | 1.45 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr2_-_131001916 | 1.43 |
ENSMUST00000103188.10
ENSMUST00000133602.8 ENSMUST00000028800.12 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr1_-_171050004 | 1.42 |
ENSMUST00000147246.2
ENSMUST00000111326.8 ENSMUST00000138184.8 |
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr12_+_104998895 | 1.41 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr4_+_150321659 | 1.41 |
ENSMUST00000133839.8
|
Eno1
|
enolase 1, alpha non-neuron |
| chr13_+_9326513 | 1.41 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr6_-_125142539 | 1.41 |
ENSMUST00000183272.2
ENSMUST00000182052.8 ENSMUST00000182277.2 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr8_-_86281946 | 1.39 |
ENSMUST00000034138.7
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr18_-_67582191 | 1.39 |
ENSMUST00000025408.10
|
Afg3l2
|
AFG3-like AAA ATPase 2 |
| chr9_+_54493618 | 1.35 |
ENSMUST00000217484.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr7_-_79115915 | 1.35 |
ENSMUST00000073889.14
|
Polg
|
polymerase (DNA directed), gamma |
| chr13_-_53083494 | 1.35 |
ENSMUST00000123599.8
|
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr14_+_8348779 | 1.33 |
ENSMUST00000022256.5
|
Psmd6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr4_+_99786611 | 1.33 |
ENSMUST00000058351.16
|
Pgm1
|
phosphoglucomutase 1 |
| chr10_-_126866658 | 1.33 |
ENSMUST00000120547.2
ENSMUST00000152054.8 |
Tsfm
|
Ts translation elongation factor, mitochondrial |
| chr7_-_126302315 | 1.30 |
ENSMUST00000173108.8
ENSMUST00000205515.2 |
Coro1a
|
coronin, actin binding protein 1A |
| chr3_-_153617782 | 1.28 |
ENSMUST00000167111.6
|
Rabggtb
|
Rab geranylgeranyl transferase, b subunit |
| chr13_-_30170031 | 1.28 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr11_+_4186391 | 1.28 |
ENSMUST00000075221.3
|
Osm
|
oncostatin M |
| chr19_+_36325683 | 1.27 |
ENSMUST00000225920.2
|
Pcgf5
|
polycomb group ring finger 5 |
| chr5_-_88823472 | 1.26 |
ENSMUST00000113234.8
ENSMUST00000153565.8 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr11_+_98632631 | 1.25 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr17_-_33879224 | 1.23 |
ENSMUST00000130946.8
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr7_-_79115760 | 1.22 |
ENSMUST00000125562.2
|
Polg
|
polymerase (DNA directed), gamma |
| chr19_+_6450641 | 1.21 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr7_-_126398165 | 1.19 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr5_+_34494272 | 1.18 |
ENSMUST00000182047.2
|
Rnf4
|
ring finger protein 4 |
| chr11_+_98932062 | 1.18 |
ENSMUST00000017637.13
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr17_-_23990479 | 1.17 |
ENSMUST00000086325.13
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr3_+_32583602 | 1.16 |
ENSMUST00000091257.11
|
Mfn1
|
mitofusin 1 |
| chr5_+_137628377 | 1.14 |
ENSMUST00000175968.8
|
Lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr5_-_31205339 | 1.13 |
ENSMUST00000202520.4
ENSMUST00000202556.4 |
Slc5a6
|
solute carrier family 5 (sodium-dependent vitamin transporter), member 6 |
| chr13_-_98152768 | 1.13 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr15_+_88746380 | 1.12 |
ENSMUST00000042818.11
|
Pim3
|
proviral integration site 3 |
| chrX_+_68403900 | 1.11 |
ENSMUST00000033532.7
|
Aff2
|
AF4/FMR2 family, member 2 |
| chr1_-_175453117 | 1.10 |
ENSMUST00000027810.14
|
Fh1
|
fumarate hydratase 1 |
| chr1_-_171050077 | 1.10 |
ENSMUST00000005817.9
|
Tomm40l
|
translocase of outer mitochondrial membrane 40-like |
| chr5_-_88823049 | 1.08 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr6_-_115739284 | 1.08 |
ENSMUST00000166254.7
ENSMUST00000170625.8 |
Tmem40
|
transmembrane protein 40 |
| chr19_-_6973393 | 1.08 |
ENSMUST00000041686.10
ENSMUST00000180765.2 |
Nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr11_+_120375439 | 1.08 |
ENSMUST00000043627.8
|
Mrpl12
|
mitochondrial ribosomal protein L12 |
| chr7_-_33929667 | 1.08 |
ENSMUST00000206415.2
|
Gpi1
|
glucose-6-phosphate isomerase 1 |
| chr19_-_32038838 | 1.07 |
ENSMUST00000096119.5
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr14_+_32043944 | 1.07 |
ENSMUST00000022480.8
ENSMUST00000228529.2 |
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr3_-_83947416 | 1.07 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chrX_-_74174450 | 1.06 |
ENSMUST00000114092.8
ENSMUST00000132501.8 ENSMUST00000153318.8 ENSMUST00000155742.2 |
Mpp1
|
membrane protein, palmitoylated |
| chr4_+_137977714 | 1.06 |
ENSMUST00000105824.8
ENSMUST00000124239.8 ENSMUST00000105823.2 ENSMUST00000105818.8 |
Sh2d5
Kif17
|
SH2 domain containing 5 kinesin family member 17 |
| chr7_-_16761732 | 1.04 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr3_+_121243395 | 1.04 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr16_-_35891739 | 1.04 |
ENSMUST00000231351.2
ENSMUST00000004057.9 |
Fam162a
|
family with sequence similarity 162, member A |
| chr11_+_3939924 | 1.03 |
ENSMUST00000109981.2
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr10_-_80691009 | 1.03 |
ENSMUST00000220225.2
ENSMUST00000035775.9 |
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_+_51447613 | 1.01 |
ENSMUST00000114445.8
ENSMUST00000114446.8 ENSMUST00000141711.3 |
Cbx3
|
chromobox 3 |
| chr7_-_130148984 | 1.01 |
ENSMUST00000160289.9
|
Nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr7_+_28050077 | 1.00 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr2_-_25911544 | 1.00 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr1_-_131025562 | 1.00 |
ENSMUST00000016672.11
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr7_-_143102792 | 0.99 |
ENSMUST00000072727.7
ENSMUST00000207948.2 ENSMUST00000208190.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr18_-_35087355 | 0.99 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr17_-_35077089 | 0.98 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr5_-_31448370 | 0.98 |
ENSMUST00000041565.11
ENSMUST00000201809.2 |
Ift172
|
intraflagellar transport 172 |
| chr19_-_6885657 | 0.98 |
ENSMUST00000149261.8
|
Prdx5
|
peroxiredoxin 5 |
| chr3_+_106020545 | 0.97 |
ENSMUST00000079132.12
ENSMUST00000139086.2 |
Chia1
|
chitinase, acidic 1 |
| chrX_-_70536198 | 0.97 |
ENSMUST00000080035.11
|
Cd99l2
|
CD99 antigen-like 2 |
| chr1_-_164285914 | 0.96 |
ENSMUST00000027863.13
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr16_+_22926504 | 0.95 |
ENSMUST00000187168.7
ENSMUST00000232287.2 ENSMUST00000077605.12 |
Eif4a2
|
eukaryotic translation initiation factor 4A2 |
| chr8_+_95564949 | 0.95 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr19_-_60862964 | 0.95 |
ENSMUST00000025961.7
|
Prdx3
|
peroxiredoxin 3 |
| chr5_+_91079068 | 0.95 |
ENSMUST00000202781.2
ENSMUST00000071652.6 |
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr16_+_43960183 | 0.93 |
ENSMUST00000159514.8
ENSMUST00000161326.8 ENSMUST00000063520.15 ENSMUST00000063542.8 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr14_-_20844074 | 0.92 |
ENSMUST00000080440.14
ENSMUST00000100837.11 ENSMUST00000071816.7 |
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr16_-_43959559 | 0.92 |
ENSMUST00000063661.13
ENSMUST00000114666.9 |
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr17_-_29566774 | 0.92 |
ENSMUST00000095427.12
ENSMUST00000118366.9 |
Mtch1
|
mitochondrial carrier 1 |
| chrX_+_20529137 | 0.92 |
ENSMUST00000001989.9
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr15_+_78314251 | 0.91 |
ENSMUST00000229622.2
ENSMUST00000162808.2 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr2_-_155771938 | 0.91 |
ENSMUST00000152766.8
ENSMUST00000139232.8 ENSMUST00000109632.8 ENSMUST00000006036.13 ENSMUST00000142655.2 ENSMUST00000159238.2 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr6_+_29853745 | 0.90 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr11_-_102298141 | 0.90 |
ENSMUST00000149777.8
ENSMUST00000154001.8 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr8_+_120588977 | 0.90 |
ENSMUST00000034287.10
|
Klhl36
|
kelch-like 36 |
| chr18_-_57108405 | 0.89 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr6_+_91133647 | 0.88 |
ENSMUST00000041736.11
|
Hdac11
|
histone deacetylase 11 |
| chr5_-_24650164 | 0.87 |
ENSMUST00000115043.8
ENSMUST00000115041.2 ENSMUST00000030800.13 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr13_+_67052978 | 0.87 |
ENSMUST00000168767.9
|
Gm10767
|
predicted gene 10767 |
| chr19_+_10160249 | 0.87 |
ENSMUST00000010807.6
|
Fads1
|
fatty acid desaturase 1 |
| chr15_+_79578141 | 0.87 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chrX_-_70536449 | 0.87 |
ENSMUST00000037391.12
ENSMUST00000114586.9 ENSMUST00000114587.3 |
Cd99l2
|
CD99 antigen-like 2 |
| chr7_-_143102524 | 0.86 |
ENSMUST00000208093.2
ENSMUST00000209098.2 |
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr19_+_37184927 | 0.86 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr10_+_79552421 | 0.86 |
ENSMUST00000099513.8
ENSMUST00000020581.3 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr13_-_104246084 | 0.85 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chr17_+_28259749 | 0.84 |
ENSMUST00000233869.2
|
Anks1
|
ankyrin repeat and SAM domain containing 1 |
| chr2_-_7400690 | 0.84 |
ENSMUST00000182404.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr7_-_4448631 | 0.84 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr19_-_7218363 | 0.83 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr19_+_10160283 | 0.83 |
ENSMUST00000235160.2
|
Fads1
|
fatty acid desaturase 1 |
| chr9_-_106125055 | 0.83 |
ENSMUST00000074082.13
|
Alas1
|
aminolevulinic acid synthase 1 |
| chr3_+_63883527 | 0.82 |
ENSMUST00000029405.8
|
Gmps
|
guanine monophosphate synthetase |
| chr11_+_76297969 | 0.82 |
ENSMUST00000021203.7
ENSMUST00000152183.2 |
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr3_-_89121147 | 0.82 |
ENSMUST00000173477.8
ENSMUST00000119222.9 |
Mtx1
|
metaxin 1 |
| chr2_-_60793536 | 0.81 |
ENSMUST00000028347.13
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr1_-_4855894 | 0.81 |
ENSMUST00000130201.8
ENSMUST00000156816.7 ENSMUST00000146665.3 |
Mrpl15
|
mitochondrial ribosomal protein L15 |
| chr6_-_52217821 | 0.80 |
ENSMUST00000121043.2
|
Hoxa10
|
homeobox A10 |
| chr17_+_85265420 | 0.79 |
ENSMUST00000080217.14
ENSMUST00000112304.10 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr19_-_41373526 | 0.79 |
ENSMUST00000059672.9
|
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr17_-_27842237 | 0.79 |
ENSMUST00000062397.13
ENSMUST00000176876.8 ENSMUST00000146321.3 |
Nudt3
|
nudix (nucleotide diphosphate linked moiety X)-type motif 3 |
| chr3_-_32791296 | 0.78 |
ENSMUST00000043966.8
|
Mrpl47
|
mitochondrial ribosomal protein L47 |
| chr6_-_124791259 | 0.78 |
ENSMUST00000172132.10
ENSMUST00000239432.2 |
Tpi1
|
triosephosphate isomerase 1 |
| chr16_-_91728531 | 0.77 |
ENSMUST00000023677.10
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr13_+_51254852 | 0.77 |
ENSMUST00000095797.6
|
Spin1
|
spindlin 1 |
| chr3_+_32583681 | 0.77 |
ENSMUST00000147350.8
|
Mfn1
|
mitofusin 1 |
| chr7_-_24672055 | 0.77 |
ENSMUST00000205871.2
|
Rabac1
|
Rab acceptor 1 (prenylated) |
| chr7_+_126575781 | 0.76 |
ENSMUST00000206450.2
ENSMUST00000205830.2 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr7_-_78432774 | 0.76 |
ENSMUST00000032841.7
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr13_+_73476629 | 0.76 |
ENSMUST00000221730.2
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr9_+_48966868 | 0.75 |
ENSMUST00000034803.10
|
Zw10
|
zw10 kinetochore protein |
| chr4_+_129407374 | 0.75 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr14_+_30973407 | 0.75 |
ENSMUST00000022458.11
|
Bap1
|
Brca1 associated protein 1 |
| chr8_-_121394742 | 0.75 |
ENSMUST00000181334.2
ENSMUST00000181950.2 ENSMUST00000181333.2 |
Emc8
Gm27021
|
ER membrane protein complex subunit 8 predicted gene, 27021 |
| chr7_+_81508741 | 0.74 |
ENSMUST00000041890.8
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr13_-_30168374 | 0.74 |
ENSMUST00000221536.2
ENSMUST00000222730.2 |
E2f3
|
E2F transcription factor 3 |
| chr8_+_84379298 | 0.74 |
ENSMUST00000019577.10
ENSMUST00000211985.2 ENSMUST00000212463.2 |
Gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr11_-_68864666 | 0.74 |
ENSMUST00000038644.5
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr6_+_85428464 | 0.73 |
ENSMUST00000032078.9
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta) |
| chr2_-_119493237 | 0.73 |
ENSMUST00000028768.2
ENSMUST00000110801.8 ENSMUST00000110802.8 |
Ndufaf1
|
NADH:ubiquinone oxidoreductase complex assembly factor 1 |
| chr12_+_85645801 | 0.73 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr11_-_113456568 | 0.73 |
ENSMUST00000071539.10
ENSMUST00000106633.10 ENSMUST00000042657.16 ENSMUST00000149034.8 |
Slc39a11
|
solute carrier family 39 (metal ion transporter), member 11 |
| chr7_-_126074222 | 0.72 |
ENSMUST00000205497.2
|
Sh2b1
|
SH2B adaptor protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrb_Esrra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 1.1 | 6.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.0 | 4.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.9 | 4.7 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.9 | 2.6 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.8 | 2.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.7 | 2.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.7 | 6.9 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.6 | 4.5 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.6 | 1.9 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.5 | 1.6 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.5 | 6.1 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.5 | 2.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.5 | 4.5 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.5 | 1.5 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.5 | 1.5 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.5 | 4.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.5 | 7.7 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.5 | 1.9 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.5 | 1.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.4 | 0.4 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.4 | 1.9 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.4 | 5.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 2.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.4 | 1.1 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.4 | 6.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.4 | 2.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.4 | 1.4 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.4 | 1.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.3 | 2.1 | GO:0061198 | fungiform papilla formation(GO:0061198) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 3.7 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.3 | 2.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 | 0.6 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.3 | 1.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.3 | 1.4 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.3 | 1.1 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.3 | 0.8 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.3 | 1.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 7.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 0.8 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 1.0 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 1.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 0.7 | GO:0030887 | positive regulation of myeloid dendritic cell activation(GO:0030887) |
| 0.2 | 0.9 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.2 | 0.7 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.2 | 0.7 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 2.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 2.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 1.7 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 0.9 | GO:0050823 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.2 | 1.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.7 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.5 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.2 | 0.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.2 | 0.5 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.2 | 0.5 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 2.9 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 1.3 | GO:0032796 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 0.6 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 3.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 1.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.9 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 6.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 1.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 1.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.6 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 1.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.0 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.9 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.7 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 0.4 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.1 | 0.5 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.4 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.9 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 1.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.1 | 1.3 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.4 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.5 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.4 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.1 | 0.4 | GO:0006218 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.1 | 0.8 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.5 | GO:0015744 | succinate transport(GO:0015744) |
| 0.1 | 0.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 1.6 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.1 | 0.3 | GO:0002660 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.1 | 0.8 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.4 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.3 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 0.3 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.9 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 2.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.5 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.6 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.8 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.8 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 1.3 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 1.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.0 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 2.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 0.7 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 1.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 3.3 | GO:0071353 | ribosomal large subunit assembly(GO:0000027) cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.5 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.1 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.1 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.7 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.2 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.1 | 0.7 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.8 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 6.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 1.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.9 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.6 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.5 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 1.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.1 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 1.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 1.6 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.7 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.1 | 0.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.2 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 2.0 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.8 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.4 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.1 | 2.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.5 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.7 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 2.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.7 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.3 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.9 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.7 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.5 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 1.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:0032827 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.2 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.4 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 1.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.8 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.5 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 1.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.4 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.1 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.2 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 2.1 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.5 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.9 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.8 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.8 | GO:0007143 | female meiotic division(GO:0007143) rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0032899 | regulation of neurotrophin production(GO:0032899) |
| 0.0 | 0.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 1.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.9 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 5.2 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.5 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.7 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.9 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 1.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.6 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.7 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 1.8 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 2.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.9 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.2 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.4 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.5 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.8 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.8 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.2 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 1.7 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.0 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 0.0 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 1.0 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.0 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.5 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 2.2 | GO:0071229 | cellular response to acid chemical(GO:0071229) |
| 0.0 | 0.1 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.0 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.9 | 2.6 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.8 | 2.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.7 | 7.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.7 | 6.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.6 | 7.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.6 | 9.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.4 | 3.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.4 | 1.3 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.4 | 6.4 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.3 | 1.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.3 | 2.1 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.3 | 1.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 1.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.3 | 4.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.5 | GO:0034657 | GID complex(GO:0034657) |
| 0.3 | 8.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 0.9 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.3 | 1.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 3.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 0.8 | GO:1990423 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.2 | 0.7 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.2 | 2.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 1.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.2 | 3.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.1 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 1.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 8.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 2.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.4 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 2.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) symmetric synapse(GO:0032280) |
| 0.1 | 5.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 20.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.5 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.5 | GO:0022624 | proteasome regulatory particle(GO:0005838) proteasome accessory complex(GO:0022624) |
| 0.1 | 13.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 3.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 3.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 7.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 2.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 2.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.0 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.0 | 0.0 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 17.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 1.6 | 7.9 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 1.5 | 6.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 1.0 | 2.9 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.9 | 2.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.7 | 1.5 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.7 | 2.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.6 | 7.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.6 | 3.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.6 | 7.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.6 | 9.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 2.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.6 | 1.7 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.5 | 9.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 1.4 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.4 | 1.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 1.1 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.4 | 1.8 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.4 | 1.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.3 | 2.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 2.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.3 | 4.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.3 | 2.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.3 | 1.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.3 | 1.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 1.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.3 | 2.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 0.7 | GO:0032394 | MHC class Ib receptor activity(GO:0032394) |
| 0.2 | 1.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.7 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 1.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 3.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 1.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 0.5 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.2 | 0.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.2 | 0.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.5 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.2 | 4.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.7 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.2 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 0.5 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 2.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.6 | GO:0004104 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) |
| 0.2 | 1.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 0.9 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 0.5 | GO:0018169 | ribosomal S6-glutamic acid ligase activity(GO:0018169) |
| 0.2 | 9.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 1.6 | GO:0070061 | fructose binding(GO:0070061) |
| 0.1 | 3.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.4 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 0.1 | 1.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 1.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.9 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.1 | 0.9 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 0.4 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.4 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 1.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 1.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 1.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 2.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.3 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 1.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.9 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 1.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 2.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 2.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.2 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.1 | 1.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 2.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.4 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.6 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.6 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.5 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 1.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 1.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 1.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 5.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.4 | GO:0097617 | annealing activity(GO:0097617) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 2.9 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 4.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0035478 | chylomicron binding(GO:0035478) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 1.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 3.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 2.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 4.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 1.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 2.8 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 5.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 1.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.7 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 25.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 1.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 6.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 4.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 4.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.4 | 10.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 13.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 6.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 12.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 8.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 14.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 6.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 3.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 2.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 2.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 6.4 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.9 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.5 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.8 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.3 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 2.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |