Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
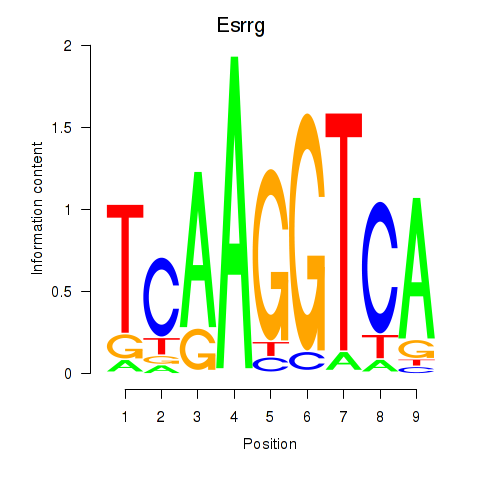
Results for Esrrg
Z-value: 1.80
Transcription factors associated with Esrrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Esrrg
|
ENSMUSG00000026610.14 | estrogen-related receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrrg | mm39_v1_chr1_+_187730018_187730031 | -0.76 | 1.4e-01 | Click! |
Activity profile of Esrrg motif
Sorted Z-values of Esrrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_32427738 | 2.22 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr2_-_73741664 | 1.99 |
ENSMUST00000111996.8
ENSMUST00000018914.3 |
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) |
| chr3_+_121243395 | 1.72 |
ENSMUST00000198393.2
|
Cnn3
|
calponin 3, acidic |
| chr10_+_127919142 | 1.51 |
ENSMUST00000026459.6
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr8_+_121395047 | 1.25 |
ENSMUST00000181795.2
|
Cox4i1
|
cytochrome c oxidase subunit 4I1 |
| chrX_+_74425990 | 1.21 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr16_+_32427789 | 1.13 |
ENSMUST00000120680.2
|
Tfrc
|
transferrin receptor |
| chr4_+_99812912 | 1.10 |
ENSMUST00000102783.5
|
Pgm1
|
phosphoglucomutase 1 |
| chr4_-_40279382 | 1.07 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr9_-_44891626 | 1.06 |
ENSMUST00000002101.12
ENSMUST00000160886.2 |
Cd3g
|
CD3 antigen, gamma polypeptide |
| chr19_+_6449776 | 1.04 |
ENSMUST00000113468.8
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr13_+_9326513 | 1.01 |
ENSMUST00000174552.8
|
Dip2c
|
disco interacting protein 2 homolog C |
| chr18_-_43610829 | 0.98 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr7_+_120234399 | 0.93 |
ENSMUST00000033176.7
ENSMUST00000208400.2 |
Uqcrc2
|
ubiquinol cytochrome c reductase core protein 2 |
| chr19_+_6449887 | 0.93 |
ENSMUST00000146601.8
ENSMUST00000150713.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr19_+_6450553 | 0.83 |
ENSMUST00000146831.8
ENSMUST00000035716.15 ENSMUST00000138555.8 ENSMUST00000167240.8 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr9_-_50515089 | 0.81 |
ENSMUST00000000175.6
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr19_-_43512929 | 0.79 |
ENSMUST00000026196.14
|
Got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr3_+_32791139 | 0.79 |
ENSMUST00000127477.8
ENSMUST00000121778.8 ENSMUST00000154257.8 |
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr2_+_32477069 | 0.78 |
ENSMUST00000102818.11
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr17_-_33879224 | 0.76 |
ENSMUST00000130946.8
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr10_+_78410803 | 0.75 |
ENSMUST00000218763.2
ENSMUST00000220430.2 ENSMUST00000218885.2 ENSMUST00000218215.2 ENSMUST00000218271.2 |
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr12_+_104998895 | 0.72 |
ENSMUST00000223244.2
ENSMUST00000021522.5 |
Glrx5
|
glutaredoxin 5 |
| chr2_+_131028861 | 0.71 |
ENSMUST00000028804.15
ENSMUST00000079857.9 |
Cdc25b
|
cell division cycle 25B |
| chr11_+_98632631 | 0.70 |
ENSMUST00000064187.12
|
Thra
|
thyroid hormone receptor alpha |
| chr9_+_107468146 | 0.66 |
ENSMUST00000195746.2
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr4_+_99786611 | 0.64 |
ENSMUST00000058351.16
|
Pgm1
|
phosphoglucomutase 1 |
| chr15_+_76227695 | 0.63 |
ENSMUST00000023210.8
ENSMUST00000231045.2 |
Cyc1
|
cytochrome c-1 |
| chr2_+_140012560 | 0.63 |
ENSMUST00000044825.5
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr17_-_26087696 | 0.62 |
ENSMUST00000236479.2
ENSMUST00000235806.2 ENSMUST00000026828.7 |
Mcrip2
|
MAPK regulated corepressor interacting protein 2 |
| chr11_-_100986192 | 0.59 |
ENSMUST00000019447.15
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr16_-_91728162 | 0.59 |
ENSMUST00000139277.8
ENSMUST00000154661.8 |
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr7_-_16761732 | 0.58 |
ENSMUST00000142597.2
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr6_-_125142539 | 0.58 |
ENSMUST00000183272.2
ENSMUST00000182052.8 ENSMUST00000182277.2 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr7_-_100232276 | 0.57 |
ENSMUST00000152876.3
ENSMUST00000150042.8 ENSMUST00000132888.9 |
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr19_+_41900360 | 0.55 |
ENSMUST00000011896.8
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr8_+_85696695 | 0.55 |
ENSMUST00000164807.2
|
Prdx2
|
peroxiredoxin 2 |
| chr15_-_35938155 | 0.55 |
ENSMUST00000156915.3
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chrX_-_47602395 | 0.54 |
ENSMUST00000114945.9
ENSMUST00000037349.8 |
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr8_+_85696453 | 0.54 |
ENSMUST00000125893.8
|
Prdx2
|
peroxiredoxin 2 |
| chr7_-_126398165 | 0.53 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr9_-_106768601 | 0.52 |
ENSMUST00000069036.14
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr8_-_95564881 | 0.52 |
ENSMUST00000034233.15
ENSMUST00000162538.9 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr9_+_54493784 | 0.52 |
ENSMUST00000167866.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr17_+_29251602 | 0.49 |
ENSMUST00000130216.3
|
Srsf3
|
serine and arginine-rich splicing factor 3 |
| chr17_-_35077089 | 0.49 |
ENSMUST00000153400.8
|
Cfb
|
complement factor B |
| chr19_+_6450641 | 0.49 |
ENSMUST00000113467.2
|
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr16_+_4501934 | 0.48 |
ENSMUST00000060067.12
ENSMUST00000115854.4 ENSMUST00000229529.2 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr12_+_85645801 | 0.48 |
ENSMUST00000177587.9
|
Jdp2
|
Jun dimerization protein 2 |
| chr2_-_25911544 | 0.48 |
ENSMUST00000136750.3
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr9_-_121686601 | 0.48 |
ENSMUST00000213124.2
ENSMUST00000215300.2 ENSMUST00000213147.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr5_-_92496730 | 0.47 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr18_-_35087355 | 0.47 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr8_+_85696396 | 0.47 |
ENSMUST00000109733.8
|
Prdx2
|
peroxiredoxin 2 |
| chr18_-_67582191 | 0.46 |
ENSMUST00000025408.10
|
Afg3l2
|
AFG3-like AAA ATPase 2 |
| chr5_-_88823472 | 0.45 |
ENSMUST00000113234.8
ENSMUST00000153565.8 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr9_-_15212849 | 0.44 |
ENSMUST00000034414.10
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr3_-_83947416 | 0.44 |
ENSMUST00000192095.6
ENSMUST00000191758.6 ENSMUST00000052342.9 |
Tmem131l
|
transmembrane 131 like |
| chr6_+_29853745 | 0.42 |
ENSMUST00000064872.13
ENSMUST00000152581.8 ENSMUST00000176265.8 ENSMUST00000154079.8 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr11_+_4186391 | 0.42 |
ENSMUST00000075221.3
|
Osm
|
oncostatin M |
| chr9_+_54493618 | 0.42 |
ENSMUST00000217484.2
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr7_-_4448631 | 0.42 |
ENSMUST00000008579.14
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr18_-_36877571 | 0.41 |
ENSMUST00000014438.5
|
Ndufa2
|
NADH:ubiquinone oxidoreductase subunit A2 |
| chr4_-_45108038 | 0.41 |
ENSMUST00000107809.9
ENSMUST00000107808.3 ENSMUST00000107807.2 ENSMUST00000107810.3 |
Tomm5
|
translocase of outer mitochondrial membrane 5 |
| chr19_+_37184927 | 0.40 |
ENSMUST00000024078.15
ENSMUST00000112391.8 |
Marchf5
|
membrane associated ring-CH-type finger 5 |
| chr12_+_85646162 | 0.40 |
ENSMUST00000050687.14
|
Jdp2
|
Jun dimerization protein 2 |
| chr11_-_115590318 | 0.39 |
ENSMUST00000106497.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chrX_-_135877607 | 0.38 |
ENSMUST00000127404.8
ENSMUST00000113071.2 |
Tmsb15l
Tmsb15b1
|
thymosin beta 15b like thymosin beta 15b1 |
| chr5_-_31448370 | 0.37 |
ENSMUST00000041565.11
ENSMUST00000201809.2 |
Ift172
|
intraflagellar transport 172 |
| chr1_-_175453117 | 0.36 |
ENSMUST00000027810.14
|
Fh1
|
fumarate hydratase 1 |
| chr17_+_85265420 | 0.35 |
ENSMUST00000080217.14
ENSMUST00000112304.10 |
Ppm1b
|
protein phosphatase 1B, magnesium dependent, beta isoform |
| chr14_+_79086665 | 0.35 |
ENSMUST00000227255.2
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr15_+_78314251 | 0.35 |
ENSMUST00000229622.2
ENSMUST00000162808.2 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr5_-_24650164 | 0.34 |
ENSMUST00000115043.8
ENSMUST00000115041.2 ENSMUST00000030800.13 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr9_+_108368032 | 0.34 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr4_+_138161958 | 0.33 |
ENSMUST00000044058.11
ENSMUST00000105813.8 ENSMUST00000105815.2 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr11_+_76297969 | 0.33 |
ENSMUST00000021203.7
ENSMUST00000152183.2 |
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr2_-_155771938 | 0.32 |
ENSMUST00000152766.8
ENSMUST00000139232.8 ENSMUST00000109632.8 ENSMUST00000006036.13 ENSMUST00000142655.2 ENSMUST00000159238.2 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr19_-_4889314 | 0.31 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr6_+_54249817 | 0.31 |
ENSMUST00000204921.3
ENSMUST00000203091.3 ENSMUST00000204115.3 ENSMUST00000203941.3 ENSMUST00000204746.2 |
Chn2
|
chimerin 2 |
| chr4_-_116228921 | 0.29 |
ENSMUST00000239239.2
ENSMUST00000239177.2 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr19_-_7218363 | 0.29 |
ENSMUST00000236769.2
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr15_-_102579463 | 0.28 |
ENSMUST00000185641.7
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) |
| chr7_+_28050077 | 0.28 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chrX_-_70536198 | 0.27 |
ENSMUST00000080035.11
|
Cd99l2
|
CD99 antigen-like 2 |
| chr2_+_109522781 | 0.27 |
ENSMUST00000111050.10
|
Bdnf
|
brain derived neurotrophic factor |
| chr11_-_115590133 | 0.27 |
ENSMUST00000106499.8
|
Grb2
|
growth factor receptor bound protein 2 |
| chrX_-_70536449 | 0.27 |
ENSMUST00000037391.12
ENSMUST00000114586.9 ENSMUST00000114587.3 |
Cd99l2
|
CD99 antigen-like 2 |
| chr5_-_121665249 | 0.26 |
ENSMUST00000152270.8
|
Mapkapk5
|
MAP kinase-activated protein kinase 5 |
| chr11_+_93935156 | 0.25 |
ENSMUST00000024979.15
|
Spag9
|
sperm associated antigen 9 |
| chr18_+_77861656 | 0.25 |
ENSMUST00000114748.2
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr14_+_79086492 | 0.25 |
ENSMUST00000040990.7
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr13_-_21685588 | 0.24 |
ENSMUST00000044043.3
|
Cox5b-ps
|
cytochrome c oxidase subunit 5B, pseudogene |
| chr1_-_16589425 | 0.24 |
ENSMUST00000159558.8
ENSMUST00000054668.13 ENSMUST00000162627.8 ENSMUST00000162007.8 ENSMUST00000128957.9 ENSMUST00000115359.10 ENSMUST00000151888.8 |
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr7_+_45224524 | 0.23 |
ENSMUST00000210811.2
|
Bcat2
|
branched chain aminotransferase 2, mitochondrial |
| chr3_+_85481416 | 0.23 |
ENSMUST00000107672.8
ENSMUST00000127348.8 ENSMUST00000107674.2 |
Gatb
|
glutamyl-tRNA(Gln) amidotransferase, subunit B |
| chr19_+_47079187 | 0.23 |
ENSMUST00000072141.4
|
Pdcd11
|
programmed cell death 11 |
| chr7_-_16761790 | 0.22 |
ENSMUST00000003183.12
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr11_-_100332619 | 0.22 |
ENSMUST00000107398.8
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr3_-_32791296 | 0.22 |
ENSMUST00000043966.8
|
Mrpl47
|
mitochondrial ribosomal protein L47 |
| chr11_-_79418500 | 0.21 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr15_+_79578141 | 0.21 |
ENSMUST00000230898.2
ENSMUST00000229046.2 |
Gtpbp1
|
GTP binding protein 1 |
| chr14_+_31807760 | 0.21 |
ENSMUST00000170600.8
ENSMUST00000168986.7 ENSMUST00000169649.2 |
Oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr19_-_6973393 | 0.20 |
ENSMUST00000041686.10
ENSMUST00000180765.2 |
Nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr8_+_95564949 | 0.20 |
ENSMUST00000034234.15
ENSMUST00000159871.4 |
Coq9
|
coenzyme Q9 |
| chr17_-_35978438 | 0.20 |
ENSMUST00000043674.15
|
Vars2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr2_-_84508385 | 0.19 |
ENSMUST00000189772.2
ENSMUST00000053664.9 ENSMUST00000111664.8 |
Gm28635
Tmx2
|
predicted gene 28635 thioredoxin-related transmembrane protein 2 |
| chr1_+_36510670 | 0.18 |
ENSMUST00000153128.2
|
Cnnm4
|
cyclin M4 |
| chr7_+_127728712 | 0.18 |
ENSMUST00000033053.8
ENSMUST00000205460.2 |
Itgax
|
integrin alpha X |
| chr9_+_108367801 | 0.17 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr17_-_26004298 | 0.17 |
ENSMUST00000150324.8
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr4_-_43584386 | 0.17 |
ENSMUST00000107884.3
|
Msmp
|
microseminoprotein, prostate associated |
| chr1_-_79838897 | 0.15 |
ENSMUST00000190724.2
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr10_-_80691009 | 0.15 |
ENSMUST00000220225.2
ENSMUST00000035775.9 |
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr4_-_82423985 | 0.14 |
ENSMUST00000107245.9
ENSMUST00000107246.2 |
Nfib
|
nuclear factor I/B |
| chr6_+_113508636 | 0.14 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr10_+_80641067 | 0.14 |
ENSMUST00000036016.6
|
Amh
|
anti-Mullerian hormone |
| chr2_-_25911691 | 0.13 |
ENSMUST00000036509.14
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr4_-_128856213 | 0.13 |
ENSMUST00000119354.8
ENSMUST00000106068.8 ENSMUST00000030581.10 |
Azin2
|
antizyme inhibitor 2 |
| chr17_-_27247581 | 0.13 |
ENSMUST00000143158.3
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr2_+_74656145 | 0.13 |
ENSMUST00000028511.8
|
Mtx2
|
metaxin 2 |
| chr19_-_4889284 | 0.13 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr10_+_79552421 | 0.13 |
ENSMUST00000099513.8
ENSMUST00000020581.3 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr7_-_115837034 | 0.12 |
ENSMUST00000182834.8
|
Plekha7
|
pleckstrin homology domain containing, family A member 7 |
| chr19_-_5738177 | 0.12 |
ENSMUST00000068169.12
|
Pcnx3
|
pecanex homolog 3 |
| chr9_-_15212745 | 0.12 |
ENSMUST00000217042.2
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr2_+_158636727 | 0.12 |
ENSMUST00000029186.14
ENSMUST00000109478.9 ENSMUST00000156893.2 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr5_+_145141333 | 0.12 |
ENSMUST00000085671.10
ENSMUST00000031601.8 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr10_-_43416941 | 0.12 |
ENSMUST00000147196.3
|
Mtres1
|
mitochondrial transcription rescue factor 1 |
| chr11_-_102298141 | 0.12 |
ENSMUST00000149777.8
ENSMUST00000154001.8 |
Slc25a39
|
solute carrier family 25, member 39 |
| chr17_-_32607859 | 0.11 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr4_-_129121676 | 0.11 |
ENSMUST00000106051.8
|
C77080
|
expressed sequence C77080 |
| chr11_+_121593582 | 0.11 |
ENSMUST00000125580.2
|
Metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr6_-_113508536 | 0.11 |
ENSMUST00000032425.7
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr5_+_122239030 | 0.11 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr17_+_6926452 | 0.10 |
ENSMUST00000097430.10
|
Sytl3
|
synaptotagmin-like 3 |
| chr19_-_37153436 | 0.09 |
ENSMUST00000142973.2
ENSMUST00000154376.8 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr15_-_42540363 | 0.08 |
ENSMUST00000022921.7
|
Angpt1
|
angiopoietin 1 |
| chr17_-_32639936 | 0.08 |
ENSMUST00000170392.9
ENSMUST00000237165.2 ENSMUST00000235892.2 ENSMUST00000114455.3 |
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr4_-_128503774 | 0.07 |
ENSMUST00000141040.2
ENSMUST00000147876.2 ENSMUST00000097877.9 |
Zscan20
|
zinc finger and SCAN domains 20 |
| chr11_+_70655035 | 0.07 |
ENSMUST00000060444.6
|
Zfp3
|
zinc finger protein 3 |
| chr11_+_100332709 | 0.07 |
ENSMUST00000001599.4
ENSMUST00000107395.3 |
Klhl10
|
kelch-like 10 |
| chr4_+_133097013 | 0.07 |
ENSMUST00000030669.8
|
Slc9a1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 1 |
| chr19_-_4665509 | 0.07 |
ENSMUST00000053597.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr16_-_8455525 | 0.07 |
ENSMUST00000052505.10
|
Tmem186
|
transmembrane protein 186 |
| chr19_-_4665668 | 0.06 |
ENSMUST00000113822.3
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr9_-_121686643 | 0.06 |
ENSMUST00000215007.2
ENSMUST00000216914.2 |
Higd1a
|
HIG1 domain family, member 1A |
| chr17_+_47696312 | 0.06 |
ENSMUST00000024774.14
|
Guca1b
|
guanylate cyclase activator 1B |
| chr11_-_69451012 | 0.06 |
ENSMUST00000004036.6
|
Efnb3
|
ephrin B3 |
| chr14_-_36690726 | 0.06 |
ENSMUST00000090024.11
|
Ccser2
|
coiled-coil serine rich 2 |
| chr13_-_103042554 | 0.06 |
ENSMUST00000171791.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_23990512 | 0.06 |
ENSMUST00000226460.2
|
Flywch1
|
FLYWCH-type zinc finger 1 |
| chr2_-_102903680 | 0.05 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr11_-_53321606 | 0.05 |
ENSMUST00000061326.5
ENSMUST00000109021.4 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr6_+_91134358 | 0.05 |
ENSMUST00000155007.2
|
Hdac11
|
histone deacetylase 11 |
| chr6_-_92160952 | 0.05 |
ENSMUST00000140438.2
|
Mrps25
|
mitochondrial ribosomal protein S25 |
| chr9_-_42372710 | 0.05 |
ENSMUST00000066179.14
|
Tbcel
|
tubulin folding cofactor E-like |
| chr13_-_91372072 | 0.05 |
ENSMUST00000022119.6
|
Atg10
|
autophagy related 10 |
| chr7_-_4448180 | 0.05 |
ENSMUST00000138798.2
|
Rdh13
|
retinol dehydrogenase 13 (all-trans and 9-cis) |
| chr16_+_17716480 | 0.05 |
ENSMUST00000055374.8
|
Tssk2
|
testis-specific serine kinase 2 |
| chrX_-_104918911 | 0.05 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr8_-_122379631 | 0.04 |
ENSMUST00000046386.5
|
Zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr19_-_53360197 | 0.04 |
ENSMUST00000086887.2
|
Gm10197
|
predicted gene 10197 |
| chr7_-_37472979 | 0.04 |
ENSMUST00000176534.8
|
Zfp536
|
zinc finger protein 536 |
| chr16_-_91728531 | 0.04 |
ENSMUST00000023677.10
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr19_-_43663282 | 0.04 |
ENSMUST00000046038.9
ENSMUST00000236433.2 |
Slc25a28
|
solute carrier family 25, member 28 |
| chr16_-_29363671 | 0.04 |
ENSMUST00000039090.9
|
Atp13a4
|
ATPase type 13A4 |
| chr19_-_7943365 | 0.03 |
ENSMUST00000182102.8
ENSMUST00000075619.5 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr2_-_140012447 | 0.03 |
ENSMUST00000046030.8
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr3_+_68776884 | 0.03 |
ENSMUST00000054551.3
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr5_+_107645626 | 0.02 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr4_-_126096376 | 0.02 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr13_-_103042294 | 0.02 |
ENSMUST00000167462.8
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_+_6973511 | 0.02 |
ENSMUST00000088223.7
|
Trpt1
|
tRNA phosphotransferase 1 |
| chr8_+_82582953 | 0.02 |
ENSMUST00000109851.3
|
Inpp4b
|
inositol polyphosphate-4-phosphatase, type II |
| chr18_+_45402018 | 0.02 |
ENSMUST00000183850.8
ENSMUST00000066890.14 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr14_-_36690636 | 0.02 |
ENSMUST00000067700.13
|
Ccser2
|
coiled-coil serine rich 2 |
| chr19_+_13505277 | 0.02 |
ENSMUST00000207987.3
ENSMUST00000207904.2 ENSMUST00000207093.3 |
Olfr1480
|
olfactory receptor 1480 |
| chr3_-_90421557 | 0.02 |
ENSMUST00000107340.2
ENSMUST00000060738.9 |
S100a1
|
S100 calcium binding protein A1 |
| chrX_-_71699740 | 0.01 |
ENSMUST00000055966.13
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr18_+_42186713 | 0.01 |
ENSMUST00000072008.11
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr4_+_34893772 | 0.01 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr9_-_54568950 | 0.01 |
ENSMUST00000128624.2
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr18_+_42186757 | 0.01 |
ENSMUST00000074679.4
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr2_+_177760959 | 0.01 |
ENSMUST00000108916.8
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr8_-_46747629 | 0.01 |
ENSMUST00000058636.9
|
Helt
|
helt bHLH transcription factor |
| chr16_+_26281885 | 0.01 |
ENSMUST00000161053.8
ENSMUST00000115302.2 |
Cldn16
|
claudin 16 |
| chrX_-_166907286 | 0.01 |
ENSMUST00000239138.2
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr14_-_70680882 | 0.00 |
ENSMUST00000000793.13
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr1_+_135727571 | 0.00 |
ENSMUST00000148201.8
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chrX_+_104807868 | 0.00 |
ENSMUST00000033581.4
|
Fgf16
|
fibroblast growth factor 16 |
| chr17_-_71575584 | 0.00 |
ENSMUST00000233148.2
|
Emilin2
|
elastin microfibril interfacer 2 |
| chr16_+_22769844 | 0.00 |
ENSMUST00000232422.2
|
Hrg
|
histidine-rich glycoprotein |
| chr16_+_22769822 | 0.00 |
ENSMUST00000023590.9
|
Hrg
|
histidine-rich glycoprotein |
| chr9_+_65536892 | 0.00 |
ENSMUST00000169003.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr11_+_7013422 | 0.00 |
ENSMUST00000020706.5
|
Adcy1
|
adenylate cyclase 1 |
| chr8_-_71315902 | 0.00 |
ENSMUST00000212611.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr10_-_78683747 | 0.00 |
ENSMUST00000061289.7
|
Olfr1356
|
olfactory receptor 1356 |
| chr2_-_77000878 | 0.00 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr11_+_42310557 | 0.00 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Esrrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.2 | 1.5 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 | 0.6 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.2 | 3.4 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.2 | 0.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.5 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 | 0.7 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 | 0.8 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.9 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.6 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 | 0.5 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.1 | 0.5 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.5 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 2.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.3 | GO:0061193 | taste bud development(GO:0061193) |
| 0.1 | 0.5 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.1 | 0.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.2 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 2.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.4 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.2 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 0.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 1.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.8 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.4 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 3.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.5 | GO:1900037 | negative regulation of skeletal muscle tissue development(GO:0048642) regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.9 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.5 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.1 | GO:0070228 | regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.8 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 1.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 2.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.4 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 2.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 1.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.9 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 1.8 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 1.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 0.8 | GO:0080130 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.3 | 0.8 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 0.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 0.6 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 0.7 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.2 | 2.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.5 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.1 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.8 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.7 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 5.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 3.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 5.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |